Abstract
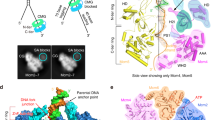
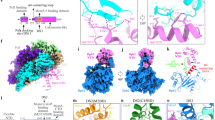
The CMG helicase is composed of Cdc45, Mcm2–7 and GINS. Here we report the structure of the Saccharomyces cerevisiae CMG, determined by cryo-EM at a resolution of 3.7–4.8 Å. The structure reveals that GINS and Cdc45 scaffold the N tier of the helicase while enabling motion of the AAA+ C tier. CMG exists in two alternating conformations, compact and extended, thus suggesting that the helicase moves like an inchworm. The N-terminal regions of Mcm2–7, braced by Cdc45–GINS, form a rigid platform upon which the AAA+ C domains make longitudinal motions, nodding up and down like an oil-rig pumpjack attached to a stable platform. The Mcm ring is remodeled in CMG relative to the inactive Mcm2–7 double hexamer. The Mcm5 winged-helix domain is inserted into the central channel, thus blocking entry of double-stranded DNA and supporting a steric-exclusion DNA-unwinding model.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Evrin, C. et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA 106, 20240–20245 (2009).
Remus, D. et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell 139, 719–730 (2009).
Botchan, M. & Berger, J. DNA replication: making two forks from one prereplication complex. Mol. Cell 40, 860–861 (2010).
Yeeles, J.T., Deegan, T.D., Janska, A., Early, A. & Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519, 431–435 (2015).
Moyer, S.E., Lewis, P.W. & Botchan, M.R. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. USA 103, 10236–10241 (2006).
Ilves, I., Petojevic, T., Pesavento, J.J. & Botchan, M.R. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 (2010).
Makarova, K.S., Koonin, E.V. & Kelman, Z. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol. Direct 7, 7 (2012).
Slaymaker, I.M. & Chen, X.S. MCM structure and mechanics: what we have learned from archaeal MCM. Subcell. Biochem. 62, 89–111 (2012).
Brewster, A.S. et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. USA 105, 20191–20196 (2008).
Enemark, E.J. & Joshua-Tor, L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 (2006).
Singleton, M.R., Sawaya, M.R., Ellenberger, T. & Wigley, D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600 (2000).
Miller, J.M., Arachea, B.T., Epling, L.B. & Enemark, E.J. Analysis of the crystal structure of an active MCM hexamer. eLife 3, e03433 (2014).
Li, N. et al. Structure of the eukaryotic MCM complex at 3.8 Å. Nature 524, 186–191 (2015).
Costa, A. et al. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. eLife 3, e03273 (2014).
Costa, A. et al. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18, 471–477 (2011).
Sun, J. et al. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 22, 976–982 (2015).
Lyubimov, A.Y., Strycharska, M. & Berger, J.M. The nuts and bolts of ring-translocase structure and mechanism. Curr. Opin. Struct. Biol. 21, 240–248 (2011).
Bochman, M.L. & Schwacha, A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 73, 652–683 (2009).
Itsathitphaisarn, O., Wing, R.A., Eliason, W.K., Wang, J. & Steitz, T.A. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151, 267–277 (2012).
Fu, Y.V. et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941 (2011).
Chen, B. & Frank, J. Two promising future developments of cryo-EM: capturing short-lived states and mapping a continuum of states of a macromolecule. Microscopy (Oxf.) doi:10.1093/jmicro/dfv344 (31 October 2015).
Chang, Y.P., Wang, G., Bermudez, V., Hurwitz, J. & Chen, X.S. Crystal structure of the GINS complex and functional insights into its role in DNA replication. Proc. Natl. Acad. Sci. USA 104, 12685–12690 (2007).
Choi, J.M., Lim, H.S., Kim, J.J., Song, O.K. & Cho, Y. Crystal structure of the human GINS complex. Genes Dev. 21, 1316–1321 (2007).
Kamada, K., Kubota, Y., Arata, T., Shindo, Y. & Hanaoka, F. Structure of the human GINS complex and its assembly and functional interface in replication initiation. Nat. Struct. Mol. Biol. 14, 388–396 (2007).
Froelich, C.A., Kang, S., Epling, L.B., Bell, S.P. & Enemark, E.J. A conserved MCM single-stranded DNA binding element is essential for replication initiation. eLife 3, e01993 (2014).
Sun, J. et al. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6–Cdt1–MCM2-7 bound to DNA. Nat. Struct. Mol. Biol. 20, 944–951 (2013).
Samel, S.A. et al. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 28, 1653–1666 (2014).
Petojevic, T. et al. Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement. Proc. Natl. Acad. Sci. USA 112, E249–E258 (2015).
Rothenberg, E., Trakselis, M.A., Bell, S.D. & Ha, T. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J. Biol. Chem. 282, 34229–34234 (2007).
Simon, A.C. et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 510, 293–297 (2014).
Onesti, S. & MacNeill, S.A. Structure and evolutionary origins of the CMG complex. Chromosoma 122, 47–53 (2013).
Krastanova, I. et al. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J. Biol. Chem. 287, 4121–4128 (2012).
Sanchez-Pulido, L. & Ponting, C.P. Cdc45: the missing RecJ ortholog in eukaryotes? Bioinformatics 27, 1885–1888 (2011).
Lairson, L.L., Henrissat, B., Davies, G.J. & Withers, S.G. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 (2008).
Wakamatsu, T. et al. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J. Biol. Chem. 285, 9762–9769 (2010).
Yamagata, A., Kakuta, Y., Masui, R. & Fukuyama, K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc. Natl. Acad. Sci. USA 99, 5908–5912 (2002).
Niedziela-Majka, A., Chesnik, M.A., Tomko, E.J. & Lohman, T.M. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro . J. Biol. Chem. 282, 27076–27085 (2007).
Velankar, S.S., Soultanas, P., Dillingham, M.S., Subramanya, H.S. & Wigley, D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 (1999).
Fischer, C.J., Maluf, N.K. & Lohman, T.M. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 344, 1287–1309 (2004).
Lee, J.Y. & Yang, W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360 (2006).
Rad, B. & Kowalczykowski, S.C. Efficient coupling of ATP hydrolysis to translocation by RecQ helicase. Proc. Natl. Acad. Sci. USA 109, 1443–1448 (2012).
Bjornson, K.P., Wong, I. & Lohman, T.M. ATP hydrolysis stimulates binding and release of single stranded DNA from alternating subunits of the dimeric E. coli Rep helicase: implications for ATP-driven helicase translocation. J. Mol. Biol. 263, 411–422 (1996).
Singleton, M.R., Dillingham, M.S. & Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 (2007).
Eisenmesser, E.Z. et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature 438, 117–121 (2005).
Frauenfelder, H., Sligar, S.G. & Wolynes, P.G. The energy landscapes and motions of proteins. Science 254, 1598–1603 (1991).
Henzler-Wildman, K. & Kern, D. Dynamic personalities of proteins. Nature 450, 964–972 (2007).
Lyubimov, A.Y., Costa, A., Bleichert, F., Botchan, M.R. & Berger, J.M. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc. Natl. Acad. Sci. USA 109, 11999–12004 (2012).
Bowman, G.D., O'Donnell, M. & Kuriyan, J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730 (2004).
Simonetta, K.R. et al. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell 137, 659–671 (2009).
Kelch, B.A., Makino, D.L., O'Donnell, M. & Kuriyan, J. How a DNA polymerase clamp loader opens a sliding clamp. Science 334, 1675–1680 (2011).
Skordalakes, E. & Berger, J.M. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114, 135–146 (2003).
Moreau, M.J., McGeoch, A.T., Lowe, A.R., Itzhaki, L.S. & Bell, S.D. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell 28, 304–314 (2007).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Scheres, S.H. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 189, 114–122 (2015).
Scheres, S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Wang, R.Y. et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat. Methods 12, 335–338 (2015).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Pettersen, E.F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Afonine, P.V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
Wei, Z. et al. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J. Biol. Chem. 285, 12469–12473 (2010).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
Cryo-EM data were collected on a Titan Krios I at the Howard Hughes Medical Institute, Janelia Farm. We also collected a cryo-EM data set on an FEI Polara with a K2 detector at the University of Texas Health Science Center. We thank the staff at these facilities for help with data collection. We also thank L. Pellegrini (University of Cambridge) for sharing the structure of human Cdc45 before publication. This work was funded by the US National Institutes of Health (GM111472 and OD12272 to H.L. and GM115809 to M.E.O'D.) and the Howard Hughes Medical Institute (M.E.O'D.).
Author information
Authors and Affiliations
Contributions
Z.Y., L.B., J.S., R.G., M.E.O'D. and H.L. designed experiments. Z.Y., L.B., J.S., R.G. and J.L. performed experiments. Z.Y., L.B., J.S. and H.L. analyzed the data. M.E.O'D. and H.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 3D classification procedure used to derive the 3.7-Å 3D density map of the Cdc45–GINS–Mcm2–7 NTD ‘platform’.
966,957 raw particles were selected from drift corrected electron micrographs. 2D classification and sorting rejected ~ 30% of the particles. 3D classification into six groups resulted in three similar 3D maps of the expected shape from our previous 15 Å CMG structure (Sun, J. et al. Nature structural & molecular biology. 12, 976-982, 2015), and the other three maps were either partial structures or distorted. Refinement using the ~470,000 particles of the three similar 3D maps led to a 3.8 Å 3D map. However, the density of the Mcm2-7 CTD motor region was noisy and indistinct. Excluding the CTD ring, the remaining region composed of Cdc45-GINS-Mcm2-7 NTD ring had an estimated resolution of 3.7 Å, based on the gold standard Fourier shell correlation curve.
Supplementary Figure 2 Euler-angle distribution of ~470,000 particles used in refinement of the 3.7-Å 3D map, and structure validation.
(a) The Thon rings in the power spectra of a typical drift-corrected electron micrograph reached to 3.0 Å. The lower left quadrant is a calculated version using the same CTF parameters as found in the experimental image. (b) The particle orientation covers all angular space. (c) Fourier shell correlations of the atomic model with the full 3D map (black), and the correlations of the 0.1-Å randomized and refined model against half map 1 (red), and with half map 2 (green), respectively. The 3.7 Å atomic model was randomly displaced by 0.1 Å. The noise-added model was refined by one round of coordinate and one round of b-factor refinement against half map 1. The refined coordinates were used to calculate FSC with half map 1, half map 2 and the full map respectively. The similarity between these curves indicates the atomic model is not over refined.
Supplementary Figure 3 Goodness of fit between the atomic model and the 3.7-Å density map of the Cdc45–GINS–Mcm2–7 NTD platform at selected subunit-interface regions.
(a) The 3D density map in blue mesh is superimposed with the atomic model in cartoon. Seven subunit interface regions were selected and magnified views are shown for the interfaces between: (I) Mcm4 and Mcm7, (II) Mcm3 and psf3, (III) Psf2 and Sld5, (IV) Mcm5 and Cdc45, (V) Cdc45 and Mcm2, (VI) Mcm2 and Mcm 6, and (VII) Mcm6 and Mcm4. (b) Five selected a-helices from Cdc45 and GINS showing the side chain densities of some of the relatively large residues.
Supplementary Figure 4 Focused 3D classification.
Two rounds of focused 3D classification in the Mcm2-7 CTD motor ring region led to the identification of the 4.8-Å map with a tilted CTD-tier ring and the 4.7-Å 3D map with a untilted CTD-tier ring. The two red lines superimposed on the Mcm2-7 NTD-tier and CTD-tier densities highlight the distinct configuration of the two conformers.
Supplementary Figure 5 3D map and resolution estimates of CMG conformer I and conformer II.
(a) Surface rendered top, front side, and right side views of the 3D map of conformer I. (b) The gold standard Fourier shell correlation suggests a resolution of 4.8 Å at the 0.143 correlation point. (c) Color-coded resolution map of conformer I. (d) Surface rendered 3D map of conformer II. (e) Conformer II has an estimated resolution of 4.7 Å at the 0.143 correlation value. (f) Color-coded resolution map of conformer II. The early drop off of the FSC curves in (b) and (e) is reflected in the pinkish color of the resolution maps, particularly at the CTD motor region. This indicates that particles assigned to each conformer still contain a certain level of conformation heterogeneity.
Supplementary Figure 6 Superimposition of electron density and structural models for the CTD rings of the two CMG conformers.
Insert figure caption here by deleting or overwriting this text; captions may run to a second page if necessary. To ensure accurate appearance in the published version, please use the Symbol font for all symbols and Greek letters.
Supplementary Figure 7 Cross-links between Mcm5 WHD and the CTD-ring interior channel (top) and the surface charge of the CMG helicase (bottom).
(a) Back side view of CMG structure with the front Mcm2 and Mcm6 removed to better display the axial channel, as highlighted with two dashed black curves. (b) An enlarged view of the left boxed area in (a) showing the two crosslinks between K750 of the Mcm5 WHD and K477 in the Mcm3 AAA+ region and K534 in the Mcm5 AAA+ region, respectively. These crosslinks were observed in the complex of CMG helicase and the leading strand polymerase e. (c) The Mcm5 WHD structure is superimposed on the electron density in the region marked by the right box in (a). The residue that crosslinks with the CTD AAA+ ring interior channel is shown in red stick representation. (d) The CTD top view. The dashed turquoise circle marks the positively charged DNA entrance. (e) CMG side view. Two dashed turquoise lines mark a narrow positive band on the outside surface of Mcm3. (f-g) Cut-open back side and front side views of CMG surface charge. The dashed turquoise squares mark the positively charged interior channel surface of the OB fold region in the NTD ring. Dashed yellow lines mark the possible DNA path inside the channel.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Table 1 (PDF 2745 kb)
Morphing of the CMG structure between conformers I and II
The video starts with an N-face view of conformer I, rotates it 90o to a back side view, then rotates it 90o to a front side view and morphs conformer I with conformer II. Then CMG conformer I is rotated 90o to a side view in which the GINSCdc45 project toward the viewer, and morphs it with conformer II. (MOV 24867 kb)
Morphing of the Mcm 5,2,6,4 subunits of CMG, showing major changes between conformers I and II
The Mcm5,2,6,4 subunits are colored yellow, blue, red, and green, respectively, and other subunits of CMG are removed for clarity. (MOV 22371 kb)
Animation of pumpjack translocation
An animation illustrating the proposed DNA unwinding and translocation mechanism of the CMG helicase, showing the side view of a ratcheting CMG acting as a nodding pumpjack while traveling from left to right on a horizontal DNA. (MOV 16979 kb)
Rights and permissions
About this article
Cite this article
Yuan, Z., Bai, L., Sun, J. et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol 23, 217–224 (2016). https://doi.org/10.1038/nsmb.3170
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3170
This article is cited by
-
Coordination of cohesin and DNA replication observed with purified proteins
Nature (2024)
-
Synergism between CMG helicase and leading strand DNA polymerase at replication fork
Nature Communications (2023)
-
Characterization of the dimeric CMG/pre-initiation complex and its transition into DNA replication forks
Cellular and Molecular Life Sciences (2020)
-
DNA translocation mechanism of the MCM complex and implications for replication initiation
Nature Communications (2019)
-
The mechanism of DNA unwinding by the eukaryotic replicative helicase
Nature Communications (2019)