Abstract
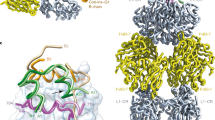
Insulins in the venom of certain fish-hunting cone snails facilitate prey capture by rapidly inducing hypoglycemic shock. One such insulin, Conus geographus G1 (Con-Ins G1), is the smallest known insulin found in nature and lacks the C-terminal segment of the B chain that, in human insulin, mediates engagement of the insulin receptor and assembly of the hormone's hexameric storage form. Removal of this segment (residues B23–B30) in human insulin results in substantial loss of receptor affinity. Here, we found that Con-Ins G1 is monomeric, strongly binds the human insulin receptor and activates receptor signaling. Con-Ins G1 thus is a naturally occurring B-chain-minimized mimetic of human insulin. Our crystal structure of Con-Ins G1 reveals a tertiary structure highly similar to that of human insulin and indicates how Con-Ins G1's lack of an equivalent to the key receptor-engaging residue PheB24 is mitigated. These findings may facilitate efforts to design ultrarapid-acting therapeutic insulins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
King, G.F. Venoms as a platform for human drugs: translating toxins into therapeutics. Expert Opin. Biol. Ther. 11, 1469–1484 (2011).
Zambelli, V.O., Pasqualoto, K.F., Picolo, G., Chudzinski-Tavassi, A.M. & Cury, Y. Harnessing the knowledge of animal toxins to generate drugs. Pharmacol. Res. http://dx.doi.org/10.1016/j.phrs.2016.01.009 (2016).
Safavi-Hemami, H. et al. Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl. Acad. Sci. USA 112, 1743–1748 (2015).
Adams, M.J. et al. Structure of rhombohedral 2 zinc insulin crystals. Nature 224, 491–495 (1969).
Owens, D.R. New horizons: alternative routes for insulin therapy. Nat. Rev. Drug Discov. 1, 529–540 (2002).
Bao, S.J., Xie, D.L., Zhang, J.P., Chang, W.R. & Liang, D.C. Crystal structure of desheptapeptide(B24–B30)insulin at 1.6 A resolution: implications for receptor binding. Proc. Natl. Acad. Sci. USA 94, 2975–2980 (1997).
Walewska, A. et al. Integrated oxidative folding of cysteine/selenocysteine containing peptides: improving chemical synthesis of conotoxins. Angew. Chem. Int. Edn. Engl. 48, 2221–2224 (2009).
Denley, A. et al. Structural determinants for high-affinity binding of insulin-like growth factor II to insulin receptor (IR)-A, the exon 11 minus isoform of the IR. Mol. Endocrinol. 18, 2502–2512 (2004).
Timofeev, V.I. et al. X-ray investigation of gene-engineered human insulin crystallized from a solution containing polysialic acid. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 259–263 (2010).
Weiss, M.A. The structure and function of insulin: decoding the TR transition. Vitam. Horm. 80, 33–49 (2009).
Menting, J.G. et al. How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245 (2013).
Menting, J.G. et al. Protective hinge in insulin opens to enable its receptor engagement. Proc. Natl. Acad. Sci. USA 111, E3395–E3404 (2014).
Muttenthaler, M., Ramos, Y.G., Feytens, D., de Araujo, A.D. & Alewood, P.F. p-Nitrobenzyl protection for cysteine and selenocysteine: a more stable alternative to the acetamidomethyl group. Biopolymers 94, 423–432 (2010).
Smith, G.D., Pangborn, W.A. & Blessing, R.H. The structure of T6 human insulin at 1.0 A resolution. Acta Crystallogr. D Biol. Crystallogr. 59, 474–482 (2003).
Rivier, J. et al. Total synthesis and further characterization of the γ-carboxyglutamate-containing “sleeper” peptide from Conus geographus venom. Biochemistry 26, 8508–8512 (1987).
Galande, A.K., Weissleder, R. & Tung, C.-H. An effective method of on-resin disulfide bond formation in peptides. J. Comb. Chem. 7, 174–177 (2005).
Houtman, J.C. et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 (2007).
Laue, T., Shah, D., Ridgeway, T. & Pelletier, S. in Analytical Ultracentrifugation in Biochemistry and Polymer Science (eds. Harding, S., Rowe, A.J. & Horton, J.C.) 90–125 (Royal Society of Chemistry, 1992).
Dai, Q., Dong, M., Liu, Z., Prorok, M. & Castellino, F.J. Ca 2+-induced self-assembly in designed peptides with optimally spaced γ-carboxyglutamic acid residues. J. Inorg. Biochem. 105, 52–57 (2011).
Cnudde, S.E., Prorok, M., Jia, X., Castellino, F.J. & Geiger, J.H. The crystal structure of the calcium-bound con-G[Q6A] peptide reveals a novel metal-dependent helical trimer. J. Biol. Inorg. Chem. 16, 257–266 (2011).
Chen, Z. et al. Conformational changes in conantokin-G induced upon binding of calcium and magnesium as revealed by NMR structural analysis. J. Biol. Chem. 273, 16248–16258 (1998).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 47, 5.6 (2014).
Hua, Q.X. et al. Structure of a protein in a kinetic trap. Nat. Struct. Biol. 2, 129–138 (1995).
Sparrow, L.G. et al. N-linked glycans of the human insulin receptor and their distribution over the crystal structure. Proteins 71, 426–439 (2008).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Guvench, O. et al. CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 7, 3162–3180 (2011).
Best, R.B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Acknowledgements
R.S.N. acknowledges fellowship support from the National Health and Medical Research Council of Australia (NHMRC). N.A.S. acknowledges receipt of an Australian Postgraduate Award scholarship. This work was supported in part by National Institutes of Health grants GM 48677 (to B.M.O. and J.E.R., a subcontractor at Sentia Medical Sciences), by NHMRC Project Grant APP1058233 (to M.C.L.) and by the Utah Science and Technology Initiative (USTAR, to D.H.-C.C.). H.S.-H. acknowledges fellowship support from the European Commission (CONBIOS 330486). Aspects of this work were made possible through Victorian State Government Operational Infrastructure Support, the Australian Government NHMRC IRIISS and a pilot grant from the University of Utah Diabetes and Metabolism Center. We thank A. Morrione (Thomas Jefferson University) for providing cells. Part of this research was undertaken on the MX2 beamline at the Australian Synchrotron (Victoria, Australia).
Author information
Authors and Affiliations
Contributions
J.G.M. and M.C.L. performed crystallography; M.C.L., R.S.N., H.S.-H. and B.J.S. directed research; B.M.O., H.S.-H., R.S.N. and M.C.L. designed the study; B.E.F. and D.H.-C.C. performed experiments and analyzed data; J.G., C.M. and J.E. synthesized peptides and analyzed data; M.M.D. performed experiments; C.A.M. performed ultracentrifugation experiments, N.A.S. and B.J.S. performed computer modeling, J.E.R. supervised peptide synthesis, and M.C.L., R.S.N., H.S.-H. and B.J.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.C.L.'s and D.H.C.'s research activities are partially funded by Sanofi (Germany).
Integrated supplementary information
Supplementary Figure 1 Arrangement of Con-Ins G1 monomers around the crystallographic four-fold axis (stereo).
The four Con-Ins G1 monomers coordinate an apparent single off-axis sulfate molecules (centre), modelled with unrestrained coordinates into a relatively featureless blob (not shown) of difference electron density lying on the crystallographic four-fold axis. The ion forms part of a charge-compensated cluster comprising the amino-terminal group of GlyA1 and a side-chain carboxylate of GlaA4 from each Con-Ins G1 monomer. The Con-Ins G1 A chains are in pink and the B chains in light blue.
Supplementary Figure 2 Amino acid sequence and RP-HPLC profile of fully oxidized Con-Ins G1.
The left-hand panel shows the amino acid sequence of fully-oxidized Con-Ins G1 (top: A chain; bottom, B chain). O: hydroxyproline, γ: γ-carboxyglutamate, *: amidated C-terminus. The right-hand panel shows the RP-HPLC profile. RP-HPLC conditions are: C18 Vydac RP-HPLC column, linear gradient ranging from 15 to 45% of solvent B in 30 min with 1 mL / min flow rate monitored at 220 nm.
Supplementary Figure 3 Amino acid sequence and RP-HPLC profile of fully oxidized PTM-free sCon-Ins G1.
The left-hand panel shows the amino acid sequence of fully-oxidized PTM-free sCon-Ins G1 (top: A chain; bottom, B chain). Residues in bold indicate site of mutation; U: selenocysteine, *: amidated C-terminus. The right-hand panel shows the RP-HPLC profile. RP-HPLC conditions are: C18 Vydac RP-HPLC column, linear gradient ranging from 15 to 45% of solvent B in 30 min with 1 mL / min flow rate monitored at 220 nm.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Note (PDF 515 kb)
Rights and permissions
About this article
Cite this article
Menting, J., Gajewiak, J., MacRaild, C. et al. A minimized human insulin-receptor-binding motif revealed in a Conus geographus venom insulin. Nat Struct Mol Biol 23, 916–920 (2016). https://doi.org/10.1038/nsmb.3292
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3292
This article is cited by
-
Determinants of IGF-II influencing stability, receptor binding and activation
Scientific Reports (2022)
-
HL-7 and HL-10 Peptides Stimulate Insulin Secretion in the INS-1 Insulinoma Cell Line through Incretin-Dependent Pathway and Increasing the Glucose Uptake in L6 Myoblast
International Journal of Peptide Research and Therapeutics (2021)
-
A structurally minimized yet fully active insulin based on cone-snail venom insulin principles
Nature Structural & Molecular Biology (2020)
-
Linking neuroethology to the chemical biology of natural products: interactions between cone snails and their fish prey, a case study
Journal of Comparative Physiology A (2017)