Abstract
RING-domain E3 ligases enhance transfer of ubiquitin to substrate proteins by stabilizing the RING-bound thioester-linked E2∼ubiquitin conjugate in a defined conformation that primes the active site for nucleophilic attack. Here we report that the monomeric RING domains from the human E3 ligases Arkadia and Ark2C bind directly to free ubiquitin with an affinity comparable to that of other dedicated ubiquitin-binding domains. Further work showed that the Ark-like RING domain and the noncovalently bound ubiquitin molecule coordinately stabilize the E2-conjugated ubiquitin (donor ubiquitin) in the 'closed' conformation. Our studies identify the RING domain of Arkadia as a ubiquitin-binding domain and provide insight into a new ubiquitin-dependent mechanism used by monomeric RING domains to activate ubiquitin transfer. This study also suggests how substrates that have been monoubiquitinated could be favored for further ubiquitination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Grabbe, C., Husnjak, K. & Dikic, I. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 12, 295–307 (2011).
Deshaies, R.J. & Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009).
Metzger, M.B., Pruneda, J.N., Klevit, R.E. & Weissman, A.M. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 (2014).
Wenzel, D.M., Lissounov, A., Brzovic, P.S. & Klevit, R.E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011).
Dou, H., Buetow, L., Sibbet, G.J., Cameron, K. & Huang, D.T. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883 (2012).
Dou, H., Buetow, L., Sibbet, G.J., Cameron, K. & Huang, D.T. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat. Struct. Mol. Biol. 20, 982–986 (2013).
Plechanovová, A., Jaffray, E.G., Tatham, M.H., Naismith, J.H. & Hay, R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012).
Pruneda, J.N. et al. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 (2012).
Buetow, L. et al. Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol. Cell 58, 297–310 (2015).
Branigan, E., Plechanovová, A., Jaffray, E.G., Naismith, J.H. & Hay, R.T. Structural basis for the RING-catalyzed synthesis of K63-linked ubiquitin chains. Nat. Struct. Mol. Biol. 22, 597–602 (2015).
Budhidarmo, R., Nakatani, Y. & Day, C.L. RINGs hold the key to ubiquitin transfer. Trends Biochem. Sci. 37, 58–65 (2012).
Koinuma, D. et al. Arkadia amplifies TGF-β superfamily signalling through degradation of Smad7. EMBO J. 22, 6458–6470 (2003).
Nagano, Y. et al. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-β signaling. J. Biol. Chem. 282, 20492–20501 (2007).
Poulsen, S.L. et al. RNF111/Arkadia is a SUMO-targeted ubiquitin ligase that facilitates the DNA damage response. J. Cell Biol. 201, 797–807 (2013).
van Cuijk, L. et al. SUMO and ubiquitin-dependent XPC exchange drives nucleotide excision repair. Nat. Commun. 6, 7499 (2015).
Erker, Y. et al. Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol. Cell. Biol. 33, 2163–2177 (2013).
Sun, H. & Hunter, T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J. Biol. Chem. 287, 42071–42083 (2012).
Sun, H., Liu, Y. & Hunter, T. Multiple Arkadia/RNF111 structures coordinate its Polycomb body association and transcriptional control. Mol. Cell. Biol. 34, 2981–2995 (2014).
Sharma, V. et al. Enhancement of TGF-β signaling responses by the E3 ubiquitin ligase Arkadia provides tumor suppression in colorectal cancer. Cancer Res. 71, 6438–6449 (2011).
Briones-Orta, M.A. et al. Arkadia regulates tumor metastasis by modulation of the TGF-β pathway. Cancer Res. 73, 1800–1810 (2013).
Zhou, F. et al. Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-β signalling. Nat. Commun. 5, 3388 (2014).
Chen, H. et al. RNF111/Arkadia is regulated by DNA methylation and affects TGF-β/Smad signaling associated invasion in NSCLC cells. Lung Cancer 90, 32–40 (2015).
Mizutani, A., Koinuma, D., Seimiya, H. & Miyazono, K. The Arkadia-ESRP2 axis suppresses tumor progression: analyses in clear-cell renal cell carcinoma. Oncogene 10.1038/onc.2015.412 (2 November 2015).
Kelly, C.E. et al. Rnf165/Ark2C enhances BMP-Smad signaling to mediate motor axon extension. PLoS Biol. 11, e1001538 (2013).
Prudden, J. et al. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 (2007).
Bessarab, D.A., Mathavan, S., Jones, C.M. & Ray Dunn, N. Zebrafish Rnf111 is encoded by multiple transcripts and is required for epiboly progression and prechordal plate development. Differentiation 89, 22–30 (2015).
Mavrakis, K.J. et al. Arkadia enhances Nodal/TGF-β signaling by coupling phospho-Smad2/3 activity and turnover. PLoS Biol. 5, e67 (2007).
Chasapis, C.T., Kandias, N.G., Episkopou, V., Bentrop, D. & Spyroulias, G.A. NMR-based insights into the conformational and interaction properties of Arkadia RING-H2 E3 Ub ligase. Proteins 80, 1484–1489 (2012).
Middleton, A.J., Budhidarmo, R. & Day, C.L. Use of E2∼ubiquitin conjugates for the characterization of ubiquitin transfer by RING E3 ligases such as the inhibitor of apoptosis proteins. Methods Enzymol. 545, 243–263 (2014).
Brzovic, P.S., Lissounov, A., Christensen, D.E., Hoyt, D.W. & Klevit, R.E.A. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 (2006).
Hofmann, R.M. & Pickart, C.M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999).
DaRosa, P.A. et al. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature 517, 223–226 (2015).
Dou, H. et al. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat. Struct. Mol. Biol. 19, 184–192 (2012).
Mace, P.D. et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J. Biol. Chem. 283, 31633–31640 (2008).
Bosanac, I. et al. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol. Cell 40, 548–557 (2010).
Scott, D., Oldham, N.J., Strachan, J., Searle, M.S. & Layfield, R. Ubiquitin-binding domains: mechanisms of ubiquitin recognition and use as tools to investigate ubiquitin-modified proteomes. Proteomics 15, 844–861 (2015).
Hurley, J.H., Lee, S. & Prag, G. Ubiquitin-binding domains. Biochem. J. 399, 361–372 (2006).
Kulathu, Y. & Komander, D. Atypical ubiquitylation: the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13, 508–523 (2012).
Hospenthal, M.K., Freund, S.M. & Komander, D. Assembly, analysis and architecture of atypical ubiquitin chains. Nat. Struct. Mol. Biol. 20, 555–565 (2013).
Kaiser, S.E. et al. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 8, 691–696 (2011).
Nakatani, Y. et al. Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase. Biochem. J. 450, 629–638 (2013).
Sakata, E. et al. Crystal structure of UbcH5b∼ubiquitin intermediate: insight into the formation of the self-assembled E2∼Ub conjugates. Structure 18, 138–147 (2010).
Lu, Y., Wang, W. & Kirschner, M.W. Specificity of the anaphase-promoting complex: a single-molecule study. Science 348, 1248737 (2015).
Kelly, A., Wickliffe, K.E., Song, L., Fedrigo, I. & Rape, M. Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol. Cell 56, 232–245 (2014).
Brown, N.G. et al. Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol. Cell 56, 246–260 (2014).
Keller, S. et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 84, 5066–5073 (2012).
Smith, D.B. & Johnson, K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67, 31–40 (1988).
Dauter, Z., Dauter, M. & Dodson, E. Jolly SAD. Acta Crystallogr. D Biol. Crystallogr. 58, 494–506 (2002).
Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G.N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Joosten, R.P., Long, F., Murshudov, G.N. & Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 (2014).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Leslie, A.G.W. & Powell, H.R. Processing diffraction data with Mosflm. Evolving Methods for Macromolecular Crystallography 245, 41–51 (2007).
Pierce, M.M., Raman, C.S. & Nall, B.T. Isothermal titration calorimetry of protein-protein interactions. Methods 19, 213–221 (1999).
Zhao, H., Piszczek, G. & Schuck, P. SEDPHAT: a platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods 76, 137–148 (2015).
Acknowledgements
We thank members of C.L.D.'s laboratory for helpful discussions. We thank M. Hinds for initial analysis of NMR samples, and M. Algie and T. Kleffman for mass spectrometry. J.D.W. acknowledges the support of a University of Otago Doctoral Scholarship, and C.L.D. acknowledges the award of a University of Otago research grant to support this work. P.D.M. is supported by a Rutherford Discovery Fellowship from the New Zealand government administered by the Royal Society of New Zealand. We also thank the Australian Synchrotron for X-ray data collection and the NZ Synchrotron Group for providing travel funds.
Author information
Authors and Affiliations
Contributions
J.D.W. performed all the experiments; C.L.D. conceived the project and together with J.D.W. designed the experiments and interpreted the data; P.D.M. assisted with data analysis. J.D.W., P.D.M. and C.L.D. jointly wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
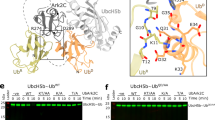
Supplementary Figure 1 Characterization of the solution properties of Ark-like RINGs.
Related to Fig. 1. (a) Sequence alignment of the RING domains from human Arkadia and Ark2C. Residues that differentiate Arkadia isoform 1 from 2 are red, zinc-coordinating residues are blue and sequence conservation is indicated. (b) ITC analysis titrating Arkadia RING (869-986, left) and Ark2C RING (228-346, right) into UbcH5b. (c) SEC-MALLS analysis of purified Arkadia RING (869-986) alone (150 μM), and in complex with UbcH5b (150 μM).
Supplementary Figure 2 Analysis of Ark-like-protein deletion constructs.
Related to Fig. 1. (a) Sequence alignments of Ark2C RING constructs with various length loop deletions. (b) GST-pulldown assay with GST-Ark2C RING variants that have different loop deletions and UbcH5b~Ub. (c) GST-pulldown assay with different GST-Arkadia isoforms and UbcH5b. (d) GST-pulldown assay with GST-Arkadia RING variants and UbcH5b or UbcH5b~Ub.
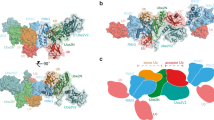
Supplementary Figure 3 Structure of Ark2C RING–UbcH5b~Ub complex 1.
Related to Fig. 2. Top, the entire asymmetric unit, consisting of four 1:1 RING–UbcH5b~Ub complexes is shown as a cartoon. RING (blue), UbcH5b (grey) and Ubiquitin (yellow) are labeled and designated with the superscripts 1–4 indicating which of the four complexes each molecule belongs to. Bottom, close up view of RING–UbR interface with electron density contoured to σ = 1.0.
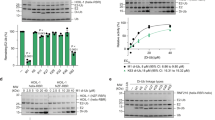
Supplementary Figure 4 Ub–RING and RING–E2 binding analysis through ITC.
Related to Fig. 3.s Top left, titrating wild-type (WT) Ub (500 μM) into WT Ark2C RING (residues 255–346) (25 μM). Top right, titrating WT Ub (500 μM) into WT Ark2C RING (20 μM) plus UbcH5bS22R (40 μM). Middle left, titrating UbcH5bS22R (250 μM) into WT Ark2C RING (20 μM). Middle right, titrating UbcH5bS22R (250 μM) into WT Ark2C RING (255-346, 20 μM) plus WT Ub (200 μM). (b) Titrating M313A Ark2C RING (residues 255–346 (250 μM) into UbcH5b (25 μM). Error bars in a,b are shown for each individual injection according to the fitting of the baseline45.
Supplementary Figure 5 Additional characterization of RING-UbR interface mutants.
Related to Fig. 3. (a) UbcH5b~Ub thioester hydrolysis assays showing the disappearance of UbcH5bS22R~Ub (15 μM) and the appearance of UbcH5b in the presence of Ark2C RING variants (2 μM) and ubiquitin variants (100 μM). (b) UbcH5b~Ub oxyester hydrolysis assays in the absence of RING, plus or minus WT ubiquitin (100 μM). (c) UbcH5b~Ub thioester hydrolysis assays showing the disappearance of UbcH5bS22R~Ub (15 μM) and the appearance of UbcH5b in the presence of WT Ark2C RING or Ub-RING fusion variants (2 μM). Samples in a,c were resolved with non-reducing SDS-PAGE. Samples in b were resolved by reducing SDS-PAGE. All samples were stained with Coomassie blue.
Supplementary Figure 6 Ubiquitin in the RING–UbcH5b~Ub complex 2 crystal structure does not adopt a canonical closed conformation.
Related to Fig. 4. (a) Overlay of the Ark2C RING–UbcH5b complexes from the complex #1 and #2 crystal structures aligned relative to UbcH5b. This showed that in complex #2 (the closed conformation structure) the RING domain had shifted slightly away from UbcH5b. (b) Cartoon representation of Ark2C RING–UbcH5b~Ub complex #2 crystal structure with a canonically closed ubiquitin (magenta) from the RNF4-UbcH5a~Ub complex structure (PDB ID: 4AP48) aligned relative to UbcH5b. (c) Close up of the linker between the ubiquitin C-terminal tail (yellow) and UbcH5b (grey) from our closed-conformation structure compared to the equivalent linker from the RNF4–UbcH5b~Ub complex structure (magenta).
Supplementary Figure 7 Predicted UbR-UbD interface mutations disrupt RING activity.
Related to Fig. 5. (a) Isothermal titration calorimetry analyses titrating G10E Ub (350 μM) into wild-type (WT) Ark2C RING (residues 255-346) (25 μM) (left) K11D Ub (600 μM) into WT Ark2C RING (30 μM) (middle) and T12D Ub (600 μM) into WT Ark2C RING (30 μM) (right). Error bars are shown for each individual injection according to the fitting of the baseline45. (b,c) UbcH5b~Ub oxyester hydrolysis assays tracking the disappearance of UbcH5bC85S,S22R~Ub and the appearance of free UbcH5b in the presence of WT Ark2C RING (5 μM) and ubiquitin variants (100 μM WT, 300 μM T12D) (b), or Ub-RING fusion variants (5 μM) (c). Top, samples were resolved with reducing SDS-PAGE and stained with Coomassie blue; experiments performed in duplicate, one set shown. Bottom, densitometry quantification from gels; error bars show range of the experimental duplicates (n=2).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 (PDF 1241 kb)
Supplementary Data Set 1
Model of the catalytic complex (PDF 14206 kb)
Supplementary Data Set 2
Uncropped gels (TXT 233 kb)
Rights and permissions
About this article
Cite this article
Wright, J., Mace, P. & Day, C. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nat Struct Mol Biol 23, 45–52 (2016). https://doi.org/10.1038/nsmb.3142
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3142
This article is cited by
-
UBE2A and UBE2B are recruited by an atypical E3 ligase module in UBR4
Nature Structural & Molecular Biology (2024)
-
The UAS thioredoxin-like domain of UBXN7 regulates E3 ubiquitin ligase activity of RNF111/Arkadia
BMC Biology (2023)
-
Structural basis for the E3 ligase activity enhancement of yeast Nse2 by SUMO-interacting motifs
Nature Communications (2021)
-
Ubiquitin transfer by a RING E3 ligase occurs from a closed E2~ubiquitin conformation
Nature Communications (2020)
-
A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases
Nature Communications (2019)