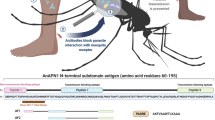
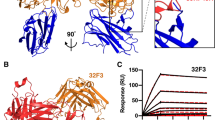
Abstract
Mosquito-based malaria transmission–blocking vaccines (mTBVs) target midgut-surface antigens of the Plasmodium parasite's obligate vector, the Anopheles mosquito. The alanyl aminopeptidase N (AnAPN1) is the leading mTBV immunogen; however, AnAPN1's role in Plasmodium infection of the mosquito and how anti-AnAPN1 antibodies functionally block parasite transmission have remained elusive. Here we present the 2.65-Å crystal structure of AnAPN1 and the immunoreactivity and transmission-blocking profiles of three monoclonal antibodies (mAbs) to AnAPN1, including mAb 4H5B7, which effectively blocks transmission of natural strains of Plasmodium falciparum. Using the AnAPN1 structure, we map the conformation-dependent 4H5B7 neoepitope to a previously uncharacterized region on domain 1 and further demonstrate that nonhuman-primate neoepitope-specific IgG also blocks parasite transmission. We discuss the prospect of a new biological function of AnAPN1 as a receptor for Plasmodium in the mosquito midgut and the implications for redesigning the AnAPN1 mTBV.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Vannice, K.S., Brown, G.V., Akanmori, B.D. & Moorthy, V.S. MALVAC 2012 scientific forum: accelerating development of second-generation malaria vaccines. Malar. J. 11, 372 (2012).
malERA Consultative Group on Vaccines. A research agenda for malaria eradication: vaccines. PLoS Med. 8, e1000398 (2011).
Sinden, R.E., Carter, R., Drakeley, C. & Leroy, D. The biology of sexual development of Plasmodium: the design and implementation of transmission-blocking strategies. Malar. J. 11, 70 (2012).
Dinglasan, R.R. & Jacobs-Lorena, M. Flipping the paradigm on malaria transmission-blocking vaccines. Trends Parasitol. 24, 364–370 (2008).
Carter, R. Transmission blocking malaria vaccines. Vaccine 19, 2309–2314 (2001).
Saul, A. Efficacy model for mosquito stage transmission blocking vaccines for malaria. Parasitology 135, 1497–1506 (2008).
Mathias, D.K. et al. Differential roles of an Anopheline midgut GPI-anchored protein in mediating Plasmodium falciparum and Plasmodium vivax ookinete invasion. Infect. Genet. Evol. 28, 635–647 (2014).
Armistead, J.S. et al. Antibodies to a single, conserved epitope in Anopheles APN1 inhibit universal transmission of Plasmodium falciparum and Plasmodium vivax malaria. Infect. Immun. 82, 818–829 (2014).
Dinglasan, R.R. et al. Disruption of Plasmodium falciparum development by antibodies against a conserved mosquito midgut antigen. Proc. Natl. Acad. Sci. USA 104, 13461–13466 (2007).
Than, M.E. et al. The endoproteinase furin contains two essential Ca2+ ions stabilizing its N-terminus and the unique S1 specificity pocket. Acta Crystallogr. D Biol. Crystallogr. 61, 505–512 (2005).
Chen, Y., Farquhar, E.R., Chance, M.R., Palczewski, K. & Kiser, P.D. Insights into substrate specificity and metal activation of mammalian tetrahedral aspartyl aminopeptidase. J. Biol. Chem. 287, 13356–13370 (2012).
McGowan, S. et al. Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase. Proc. Natl. Acad. Sci. USA 106, 2537–2542 (2009).
Kochan, G. et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. USA 108, 7745–7750 (2011).
Nguyen, T.T. et al. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat. Struct. Mol. Biol. 18, 604–613 (2011).
Luciani, N. et al. Characterization of Glu350 as a critical residue involved in the N-terminal amine binding site of aminopeptidase N (EC 3.4.11.2): insights into its mechanism of action. Biochemistry 37, 686–692 (1998).
Chen, L., Lin, Y.L., Peng, G. & Li, F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. USA 109, 17966–17971 (2012).
Wong, A.H., Zhou, D. & Rini, J.M. The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J. Biol. Chem. 287, 36804–36813 (2012).
Billker, O., Miller, A.J. & Sinden, R.E. Determination of mosquito bloodmeal pH in situ by ion-selective microelectrode measurement: implications for the regulation of malarial gametogenesis. Parasitology 120, 547–551 (2000).
Chen, J. et al. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Biochem. J. 424, 191–200 (2009).
Hua, G., Zhang, R., Abdullah, M.A.F. & Adang, M.J. Anopheles gambiae cadherin AgCad1 binds the Cry4Ba toxin of Bacillus thuringiensis israelensis and a fragment of AgCad1 synergizes toxicity. Biochemistry 47, 5101–5110 (2008).
Yaoi, K. et al. Bacillus thuringiensis Cry1Aa toxin-binding region of Bombyx mori aminopeptidase N. FEBS Lett. 463, 221–224 (1999).
Zhuang, M. et al. Heliothis virescens and Manduca sexta lipid rafts are involved in Cry1A toxin binding to the midgut epithelium and subsequent pore formation. J. Biol. Chem. 277, 13863–13872 (2002).
Nakanishi, K. et al. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella: their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 519, 215–220 (2002).
Lim, S.S. et al. Structure and dynamics of apical membrane antigen 1 from Plasmodium falciparum FVO. Biochemistry 53, 7310–7320 (2014).
Chuang, G.Y., Liou, D., Kwong, P.D. & Georgiev, I.S. NEP: web server for epitope prediction based on antibody neutralization of viral strains with diverse sequences. Nucleic Acids Res. 42, W64–W71 (2014).
Delany, I., Rappuoli, R. & De Gregorio, E. Vaccines for the 21st century. EMBO Mol. Med. 6, 708–720 (2014).
del Pilar Corena, M. et al. Carbonic anhydrase in the adult mosquito midgut. J. Exp. Biol. 208, 3263–3273 (2005).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Bahadur, R.P. & Zacharias, M. The interface of protein-protein complexes: analysis of contacts and prediction of interactions. Cell. Mol. Life Sci. 65, 1059–1072 (2008).
Yang, Y., Liu, C., Lin, Y.L. & Li, F. Structural insights into central hypertension regulation by human aminopeptidase A. J. Biol. Chem. 288, 25638–25645 (2013).
Lee, S.B., Chen, J., Aimanova, K.G. & Gill, S.S. Aedes cadherin mediates the in vivo toxicity of the Cry11Aa toxin to Aedes aegypti. Peptides 68, 140–147 (2014).
Ibrahim, M.A., Griko, N.B. & Bulla, L.A. Jr. Cytotoxicity of the Bacillus thuringiensis Cry4B toxin is mediated by the cadherin receptor BT-R(3) of Anopheles gambiae. Exp. Biol. Med. (Maywood) 238, 755–764 (2013).
Zhang, Q., Hua, G., Bayyareddy, K. & Adang, M.J. Analyses of α-amylase and α-glucosidase in the malaria vector mosquito, Anopheles gambiae, as receptors of Cry11Ba toxin of Bacillus thuringiensis subsp. jegathesan. Insect Biochem. Mol. Biol. 43, 907–915 (2013).
Hua, G., Zhang, R., Bayyareddy, K. & Adang, M.J. Anopheles gambiae alkaline phosphatase is a functional receptor of Bacillus thuringiensis jegathesan Cry11Ba toxin. Biochemistry 48, 9785–9793 (2009).
Zhang, R., Hua, G., Andacht, T.M. & Adang, M.J. A 106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae. Biochemistry 47, 11263–11272 (2008).
Likitvivatanavong, S., Chen, J., Bravo, A., Soberón, M. & Gill, S.S. Cadherin, alkaline phosphatase, and aminopeptidase N as receptors of Cry11Ba toxin from Bacillus thuringiensis subsp. jegathesan in Aedes aegypti. Appl. Environ. Microbiol. 77, 24–31 (2011).
Lal, K. et al. Characterisation of Plasmodium invasive organelles; an ookinete microneme proteome. Proteomics 9, 1142–1151 (2009).
Ecker, A., Bushell, E.S., Tewari, R. & Sinden, R.E. Reverse genetics screen identifies six proteins important for malaria development in the mosquito. Mol. Microbiol. 70, 209–220 (2008).
Ecker, A., Pinto, S.B., Baker, K.W., Kafatos, F.C. & Sinden, R.E. Plasmodium berghei: Plasmodium perforin-like protein 5 is required for mosquito midgut invasion in Anopheles stephensi. Exp. Parasitol. 116, 504–508 (2007).
Kadota, K., Ishino, T., Matsuyama, T., Chinzei, Y. & Yuda, M. Essential role of membrane-attack protein in malarial transmission to mosquito host. Proc. Natl. Acad. Sci. USA 101, 16310–16315 (2004).
Hua, G., Tsukamoto, K., Rasilo, M.L. & Ikezawa, H. Molecular cloning of a GPI-anchored aminopeptidase N from Bombyx mori midgut: a putative receptor for Bacillus thuringiensis CryIA toxin. Gene 214, 177–185 (1998).
Banks, D.J., Hua, G. & Adang, M.J. Cloning of a Heliothis virescens 110 kDa aminopeptidase N and expression in Drosophila S2 cells. Insect Biochem. Mol. Biol. 33, 499–508 (2003).
Leslie, A.W. & Powell, H. in Evolving Methods for Macromolecular Crystallography Vol. 245 (eds. Read, R. & Sussman, J.) 41–51 (Springer, 2007).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Laue, T., Shah, B., Ridgeway, T. & Pelletier, S. in Analytical Ultracentrifugation in Biochemistry and Polymer Science, (eds. Harding, S.E., Rowe, A.J. & Horton, J.C.) 90–125 (Royal Soc. Chem., Cambridge, 1992).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Brown, P.H. & Schuck, P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 90, 4651–4661 (2006).
Churcher, T.S. et al. Measuring the blockade of malaria transmission; an analysis of the standard membrane feeding assay. Int. J. Parasitol. 42, 1037–1044 (2012).
Acknowledgements
We thank the staff at the MX2 beamline of the Australian Synchrotron for assistance with X-ray data collection and the Monash Macromolecular Crystallization Facility for initial crystallization experiments. We also thank Y. Mok at the Macromolecular Interactions Facility at the University of Melbourne for assistance with sedimentation velocity experiments and J. Plieskatt at George Washington University for valuable comments and suggestions. We thank P. Eggleston and H. Hurd (both at Keele University) for the A. gambiae KEELE strain. Last, but not least, we thank the children, parents and the community of Mfou for their eager participation in this study. The work was funded in part by the PATH-Malaria Vaccine Initiative and Bloomberg Family Foundation through the Johns Hopkins Malaria Research Institute (to R.R.D.). J.S.A. was supported as a Johns Hopkins Malaria Research Institute predoctoral fellow, and D.K.M. was supported as a Calvin and Helen Lang Postdoctoral Fellow in the Biological Sciences. M.M.S. is supported by an Institut de Recherche pour le Développement Fellowship. B.B.T. is funded by the Ifakara Health Institute. S.C.A. is supported by an Australian National Health and Medical Research Council Early Career Fellowship (1072267). N.A.B. is supported by an Australian Research Council Future Fellowship (110100223). This publication was also made possible by the US National Institutes of Health National Center for Research Resources (UL1 RR 025005).
Author information
Authors and Affiliations
Contributions
S.C.A., J.S.A., D.K.M., M.M.S., D.T., N.B.-D. and I.M. designed and performed experiments. S.C.A., J.S.A., R.R.D. and N.A.B. analyzed structural and biochemical data. B.B.T., I.M. and R.R.D. analyzed all functional data. S.C.A., J.S.A., R.R.D. and N.A.B. led the manuscript preparation, with critical contributions from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Zinc and peptide within the AnAPN1 active site.
(a) Omit map for peptide in active site of molecule A, contoured at 1.8 σ. (b) Omit map for peptide in active site of molecule B, contoured at 1.6 σ. (c) Active site of AnAPN1 (molecule A) with zinc specific DANO map shown in pink, contoured at 7 σ. Key catalytic residues His366, His370 (both blue) and Glu389 (orange), as well as the active site bound peptide (green) are shown. Zinc is shown as a gray sphere.
Supplementary Figure 2 Sequence alignment of AnAPN1 and porcine APN showing the location of secondary-structure elements of AnAPN1.
Blue shading indicates positions that have fully conserved residues. The β-strands are shown as arrows and α-helices as blocks. AnAPN1 and porcine APN have moderate sequence identity (32% overall). Sequences were aligned with Clustal Omega and the alignment was edited and annotated using Jalview (version 2.8.1) and Adobe Illustrator CC.
Supplementary Figure 3 Different arrangements of domain IV from various aminopeptidases.
Surface representation of (a) AnAPN1, with a large cleft between domains II and IV, which is similar to that observed for (b) porcine APN (PDB ID 4FKE11), and (c) the open conformation of ERAP1 (pale blue, PDB ID 3MDJ14), but distinct from (d) human APN (yellow, PDB ID 4FYQ17) and (e) closed ERAP1 (bright blue, PDB ID 2YD0 (ref. 13)).
Supplementary Figure 4 Sedimentation velocity analyses of AnAPN1.
(a) Absorbance at 230 nm plotted as a function of radial position (cm) for AnAPN1 at an initial concentration of 0.15 mg/mL. Raw data (open circles) are plotted at time intervals of 8 mins and are overlaid with the continuous size-distribution best-fit (solid line) shows in (c). (b) Continuous sedimentation coefficient distribution (c(s)) plotted as a function of standardized sedimentation coefficient (s20,w) for AnAPN1 at 0.15 mg/mL. (c) Continuous mass distribution (c(M)) plotted as a function of molar mass (kDa) for AnAPN1 at 0.15 mg/mL.
Supplementary Figure 5 Epitope and functional activity analyses of monoclonal antibodies against the E. coli–expressed recombinant NT135aaAnAPN1.
(a-b) Peptide ELISA analyses of 2A12 and 4H5B7 epitope specificity. Gray bars are normal mouse IgG controls and black bars represent respective mAbs. (c) Replicate studies demonstrating the lack of transmission-blocking activity for mAbs 2A12 and 2C6 as compared to normal mouse (NM) serum IgG as measured by the Standard Membrane Feeding Assay (SMFA). (d) Immunoblot analysis of 4H5B7 mAb recognition of recombinant NT135aaAnAPN1 (double arrowhead) and near full-length AnAPN1 (single arrowhead), which was used for structural characterization. (e) The transmission-blocking activity of peptide 5-specific IgG (Pep 5 only) from NHPs that were immunized with the E. coli-expressed recombinant NT135aaAnAPN1. Peptide 5-specific IgG was depleted (P5-dep) from pooled NHP serum. (f) The transmission-blocking activity of NHP anti-Pep 7 IgG isolated by affinity chromatography (10 µg/mL) and 500 μg/mL of total IgG from mice following immunization with KLH-P7. (g-h) The transmission-blocking activity of serial dilutions of mAb 4H5B7 as compared to control mAb 2A12 (25 µg/mL) as measured by SMFA in An. stephensi NIH strain (g) and An. stephensi Nijmegen strain (h). The horizontal bars in panels c, e, g, h represent median oocyst number per mosquito midgut for each group and panel f is mean oocyst intensity. All SMFA data were analyzed statistically using a zero-inflated Generalized Linear Mixed Model as appropriate and the corresponding P-values and raw data are found in Source Data.
Supplementary Figure 6 Clustal Omega sequence alignment of APN from Bombyx mori versus AnAPN1.
Blue shading indicates positions that have fully conserved residues. The Cry1Aa toxin binding site on B. mori (NP_001037013.1) is denoted by a cyan bar above the sequences, while purple and green bars denote AnAPN1 peptides 7 and 9, respectively.
Supplementary Figure 7 Purification scheme for near-full-length recombinant AnAPN1.
(a) Chromatogram of HPLC-SEC separation of crude recombinant AnAPN1 secreted from S2 cells. (b) SDS-PAGE of HPLC fractions visualized on a Li-Cor Odyssey scanner. Peak 1 = HPLC peak at time 10.73 min (contaminating protein) and Peak 2 = HPLC peak at time 12.37 min (recombinant AnAPN1).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–3 (PDF 1118 kb)
Rights and permissions
About this article
Cite this article
Atkinson, S., Armistead, J., Mathias, D. et al. The Anopheles-midgut APN1 structure reveals a new malaria transmission–blocking vaccine epitope. Nat Struct Mol Biol 22, 532–539 (2015). https://doi.org/10.1038/nsmb.3048
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3048
This article is cited by
-
Are malaria transmission-blocking vaccines acceptable to high burden communities? Results from a mixed methods study in Bo, Sierra Leone
Malaria Journal (2021)
-
Immunofocusing humoral immunity potentiates the functional efficacy of the AnAPN1 malaria transmission-blocking vaccine antigen
npj Vaccines (2021)
-
Molecular interactions between parasite and mosquito during midgut invasion as targets to block malaria transmission
npj Vaccines (2021)
-
Anopheles metabolic proteins in malaria transmission, prevention and control: a review
Parasites & Vectors (2020)
-
Identification, molecular characterization and expression of aminopeptidase N-1 (APN-1) from Anopheles stephensi in SF9 cell line as a candidate molecule for developing a vaccine that interrupt malaria transmission
Malaria Journal (2020)