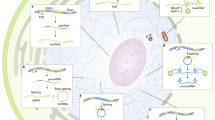
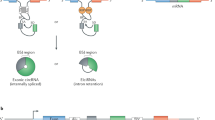
Abstract
The ribosome, central to protein synthesis in all cells, is a complex multicomponent assembly with rRNA at its functional core. During the process of ribosome biogenesis, diverse noncoding RNAs participate in controlling the quantity and quality of this rRNA. In this Review, I discuss the multiple roles assumed by noncoding RNAs during the different steps of ribosome biogenesis and how they contribute to the generation of ribosome heterogeneity, which affects normal and pathophysiological processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Voorhees, R.M. & Ramakrishnan, V. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 82, 203–236 (2013).
Lafontaine, D.L.A. 'Garbage can' for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem. Sci. 35, 267–277 (2010).
Narla, A. & Ebert, B.L. Ribosomopathies: human disorders of ribosome dysfunction. Blood 115, 3196–3205 (2010).
Freed, E.F., Bleichert, F., Dutca, L.M. & Baserga, S.J. When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst. 6, 481–493 (2010).
Gilbert, W.V. Functional specialization of ribosomes? Trends Biochem. Sci. 36, 127–132 (2011).
Xue, S. & Barna, M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369 (2012).
Ramagopal, S. Are eukaryotic ribosomes heterogeneous? Affirmations on the horizon. Biochem. Cell Biol. 70, 269–272 (1992).
Oeffinger, M. et al. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of Pre-60S ribosomal RNA. Mol. Cell 36, 768–781 (2009).
Schillewaert, S., Wacheul, L., Lhomme, F. & Lafontaine, D.L. The evolutionarily conserved protein Las1 is required for pre-rRNA processing at both ends of ITS2. Mol. Cell. Biol. 32, 430–444 (2012).
Woolford, J.L. Jr. & Baserga, S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 (2013).
Strunk, B.S. & Karbstein, K. Powering through ribosome assembly. RNA 15, 2083–2104 (2009).
Kressler, D., Hurt, E. & Bassler, J. Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 (2010).
Strunk, B.S. et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science 333, 1449–1453 (2011).Cryo-EM reconstructions of late yeast 40S precursors showing that AFs mask essential ribosomal functional areas, including the decoding site and ribosome ligand-binding sites.
Bussiere, C., Hashem, Y., Arora, S., Frank, J. & Johnson, A.W. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J. Cell Biol. 197, 747–759 (2012).
Bradatsch, B. et al. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat. Struct. Mol. Biol. 19, 1234–1241 (2012).
Matsuo, Y. et al. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 505, 112–116 (2014).
Tafforeau, L. et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell 51, 539–551 (2013). Identification of 286 human pre-rRNA processing factors, including 75 without yeast homologs. Demonstration that some human factors with yeast homologs have acquired additional processing functions.
Sloan, K.E. et al. Both endonucleolytic and exonucleolytic cleavage mediate ITS1 removal during human ribosomal RNA processing. J. Cell Biol. 200, 577–588 (2013).
Kos, M. & Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 37, 809–820 (2010).
Osheim, Y.N. et al. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16, 943–954 (2004).
Morrissey, J.P. & Tollervey, D. Birth of the snoRNPs: the evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem. Sci. 20, 78–82 (1995).
Lamaye, F., Galliot, S., Alibardi, L., Lafontaine, D.L. & Thiry, M. Nucleolar structure across evolution: the transition between bi- and tri-compartmentalized nucleoli lies within the class Reptilia. J. Struct. Biol. 174, 352–359 (2011).
Thiry, M. & Lafontaine, D.L. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 15, 194–199 (2005).
Watkins, N.J. & Bohnsack, M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 3, 397–414 (2012).
Henry, Y. et al. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13, 2452–2463 (1994).
Mullineux, S.T. & Lafontaine, D.L. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie 94, 1521–1532 (2012).
Esakova, O. & Krasilnikov, A.S. Of proteins and RNA: the RNase P/MRP family. RNA 16, 1725–1747 (2010).
Dragon, F. et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417, 967–970 (2002).
Peculis, B.A. & Steitz, J.A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73, 1233–1245 (1993).
Sharma, K. & Tollervey, D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 19, 6012–6019 (1999).
Peculis, B.A. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol. 17, 3702–3713 (1997).
Peculis, B.A. & Greer, C.L. The structure of the ITS2-proximal stem is required for pre-rRNA processing in yeast. RNA 4, 1610–1622 (1998).
Bassler, J. et al. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell 38, 712–721 (2010).Late steps of ribosome biogenesis are disassembly pathways, as illustrated here with Rea1, which 'strips' the large-subunit precursors of AFs.
Ulbrich, C. et al. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell 138, 911–922 (2009).
Kressler, D., Hurt, E., Bergler, H. & Bassler, J. The power of AAA-ATPases on the road of pre-60S ribosome maturation–molecular machines that strip pre-ribosomal particles. Biochim. Biophys. Acta 1823, 92–100 (2012).
Boulon, S., Bertrand, E. & Pradet-Balade, B. HSP90 and the R2TP co-chaperone complex: building multi-protein machineries essential for cell growth and gene expression. RNA Biol. 9, 148–154 (2012).
Darzacq, X. et al. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21, 2746–2756 (2002).
Machado-Pinilla, R., Liger, D., Leulliot, N. & Meier, U.T. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA 18, 1833–1845 (2012).
Boulon, S. et al. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 180, 579–595 (2008).
Zhao, R. et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180, 563–578 (2008).
Loc'h, J. et al. RNA mimicry by the Fap7 adenylate kinase in ribosome biogenesis. PLoS Biol. 12, e1001860 (2014).
Walbott, H. et al. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Genes Dev. 25, 2398–2408 (2011).
Li, S., Duan, J., Li, D., Ma, S. & Ye, K. Structure of the Shq1-Cbf5-Nop10-Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO J. 30, 5010–5020 (2011).
Leulliot, N. et al. The box H/ACA RNP assembly factor Naf1p contains a domain homologous to Gar1p mediating its interaction with Cbf5p. J. Mol. Biol. 371, 1338–1353 (2007).
Rothé, B. et al. Characterization of the interaction between protein Snu13p/15.5K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic Acids Res. 42, 2015–2036 (2014).
Ben-Shem, A. et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529 (2011).
Gunderson, J.H. et al. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science 238, 933–937 (1987).
Velichutina, I.V., Rogers, M.J., McCutchan, T.F. & Liebman, S.W. Chimeric rRNAs containing the GTPase centers of the developmentally regulated ribosomal rRNAs of Plasmodium falciparum are functionally distinct. RNA 4, 594–602 (1998).
Azpurua, J. et al. Naked mole-rat has increased translational fidelity compared with the mouse, as well as a unique 28S ribosomal RNA cleavage. Proc. Natl. Acad. Sci. USA 110, 17350–17355 (2013).
Birkedal, U. et al. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. Engl. doi:10.1002/anie.201408362 (21 November 2014). Mapping all rRNA 2′-O methyl groups 'at once' in yeast cells by high-throughput sequencing reveals that all positions are not fully modified at all times.
Belin, S. et al. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 4, e7147 (2009).
Buchhaupt, M. et al. Partial methylation at Am100 in 18S rRNA of baker's yeast reveals ribosome heterogeneity on the level of eukaryotic rRNA modification. PLoS ONE 9, e89640 (2014).Demonstration in yeast cells, via a DNAzyme cleavage–based assay and quantitative MS, that not all rRNA positions susceptible to 2′-O methylation are fully modified at all times.
Decatur, W.A. & Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 (2002).
Bellodi, C. et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Reports 3, 1493–1502 (2013).The catalytic activity of the pseudouridine synthase DKC1 is required for hematopoietic stem-cell differentiation.
Yoon, A. et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 312, 902–906 (2006).
Jack, K. et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44, 660–666 (2011).Yeast and mammalian ribosomes with reduced levels of pseudouridines show reduced ribosomal ligand-binding capacity, reduced IRES-dependent translation initiation and decreased translational fidelity.
Castle, J.C. et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS ONE 5, e11779 (2010).
Hughes, M.E., Grant, G.R., Paquin, C., Qian, J. & Nitabach, M.N. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 22, 1266–1281 (2012).
Jouffe, C. et al. The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11, e1001455 (2013).
Su, H. et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 33, 1348–1358 (2014).Knocking down the methyltransferase FBL in mammary cancer cells favors the IRES-dependent translation of p53.
Martens-Uzunova, E.S. et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene 31, 978–991 (2012).
Mei, Y.P. et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 31, 2794–2804 (2012).Mouse xenograft models showing that specific snoRNAs seem to have an important role in lung cancer tumorigenesis. Illustrated for breast cancer in ref. 60.
Marcel, V. et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24, 318–330 (2013).Breast and colon cancer cells deficient for p53 overexpress the methyltransferase FBL, generating hypermodified ribosomes that are deficient for translation fidelity and are more prone to initiate translation of pro-oncogenic IRES-dependent messages.
Krastev, D.B. et al. A systematic RNAi synthetic interaction screen reveals a link between p53 and snoRNP assembly. Nat. Cell Biol. 13, 809–818 (2011).
Liao, J. et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer 9, 198 (2010).
Gee, H.E. et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br. J. Cancer 104, 1168–1177 (2011).
Appaiah, H.N. et al. Persistent upregulation of U6:SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res. 13, R86 (2011).
Mannoor, K., Liao, J. & Jiang, F. Small nucleolar RNAs in cancer. Biochim. Biophys. Acta 1826, 121–128 (2012).
Houseley, J., Kotovic, K., El Hage, A. & Tollervey, D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 26, 4996–5006 (2007).
Mayer, C., Schmitz, K.M., Li, J., Grummt, I. & Santoro, R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 22, 351–361 (2006).
Schmitz, K.M., Mayer, C., Postepska, A. & Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269 (2010).
Manelyte, L., Strohner, R., Gross, T. & Langst, G. Chromatin targeting signals, nucleosome positioning mechanism and non-coding RNA-mediated regulation of the chromatin remodeling complex NoRC. PLoS Genet. 10, e1004157 (2014).
Audas, T.E., Jacob, M.D. & Lee, S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 45, 147–157 (2012).
Taft, R.J. et al. Small RNAs derived from snoRNAs. RNA 15, 1233–1240 (2009).
Scott, M.S., Avolio, F., Ono, M., Lamond, A.I. & Barton, G.J. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 5, e1000507 (2009).
Ono, M. et al. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 39, 3879–3891 (2011).
Scott, M.S. et al. Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Res. 40, 3676–3688 (2012).
Ender, C. et al. A human snoRNA with microRNA-like functions. Mol. Cell 32, 519–528 (2008).
Saraiya, A.A. & Wang, C.C. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 4, e1000224 (2008).
Rogler, L.E. et al. Small RNAs derived from lncRNA RNase MRP have gene-silencing activity relevant to human cartilage-hair hypoplasia. Hum. Mol. Genet. 23, 368–382 (2014).
Xiao, J., Lin, H., Luo, X., Luo, X. & Wang, Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 30, 524–532 (2011).
Kishore, S. & Stamm, S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311, 230–232 (2006).
Kishore, S. et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 19, 1153–1164 (2010).
Yin, Q.F. et al. Long noncoding RNAs with snoRNA ends. Mol. Cell 48, 219–230 (2012).
Tessarz, P. et al. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505, 564–568 (2014).
Jobert, L. et al. The human base excision repair enzyme SMUG1 directly interacts with DKC1 and contributes to RNA quality control. Mol. Cell 49, 339–345 (2013).
Panse, V.G. & Johnson, A.W. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35, 260–266 (2010).
Schäfer, T. et al. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 441, 651–655 (2006).Cryo-EM work in yeast showing that the protruding beak probably unfolds only once small-subunit precursors have reached the cytoplasm.
Lo, K.Y. et al. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell 39, 196–208 (2010).
Strunk, B.S., Novak, M.N., Young, C.L. & Karbstein, K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 150, 111–121 (2012).
Lebaron, S. et al. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat. Struct. Mol. Biol. 19, 744–753 (2012).
García-Gómez, J.J. et al. Final pre-40S maturation depends on the functional integrity of the 60S subunit ribosomal protein L3. PLoS Genet. 10, e1004205 (2014).
Duan, J., Li, L., Lu, J., Wang, W. & Ye, K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol. Cell 34, 427–439 (2009).
Grummt, I. Wisely chosen paths: regulation of rRNA synthesis. FEBS J. 277, 4626–4639 (2010).
Acknowledgements
S.-T. Mullineux (Agriculture and Agri-Food Canada Research Centre) is acknowledged for critical reading of the manuscript. N. Leulliot (Laboratoire de Cristallographie et RMN Biologiques, UMR CNRS 8015, Université Paris Descartes, Faculté de Pharmacie, Sorbonne Paris Cité) and K. Karbstein and H. Ghalei (Department of Cancer Biology, Scripps Research Institute) are acknowledged for assistance in figure production. The laboratory of D.L.J.L. is supported by Fonds de la Recherche Scientifique, Université Libre de Bruxelles, Center for Microscopy and Molecular Imaging (Cmmi), the European Regional Development Fund and Région Wallonne (DGO6).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Lafontaine, D. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 22, 11–19 (2015). https://doi.org/10.1038/nsmb.2939
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2939
This article is cited by
-
PQBP5/NOL10 maintains and anchors the nucleolus under physiological and osmotic stress conditions
Nature Communications (2023)
-
The nucleolus of Giardia and its ribosomal biogenesis
Parasitology Research (2023)
-
Glutamine deficiency in solid tumor cells confers resistance to ribosomal RNA synthesis inhibitors
Nature Communications (2022)
-
The nucleolus as a multiphase liquid condensate
Nature Reviews Molecular Cell Biology (2021)
-
Emerging role of non‐coding RNA in health and disease
Metabolic Brain Disease (2021)