Abstract
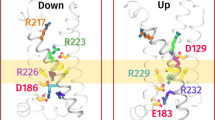
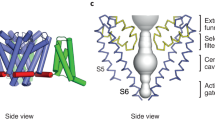
The transduction of transmembrane electric fields into protein motion has an essential role in the generation and propagation of cellular signals. Voltage-sensing domains (VSDs) carry out these functions through reorientations of positive charges in the S4 helix. Here, we determined crystal structures of the Ciona intestinalis VSD (Ci-VSD) in putatively active and resting conformations. S4 undergoes an ~5-Å displacement along its main axis, accompanied by an ~60° rotation. This movement is stabilized by an exchange in countercharge partners in helices S1 and S3 that generates an estimated net charge transfer of ~1 eo. Gating charges move relative to a ''hydrophobic gasket' that electrically divides intra- and extracellular compartments. EPR spectroscopy confirms the limited nature of S4 movement in a membrane environment. These results provide an explicit mechanism for voltage sensing and set the basis for electromechanical coupling in voltage-dependent enzymes and ion channels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
23 February 2014
In the version of this article initially published online, an incorrect affiliation was listed for Klaus Schulten. Klaus Schulten is affiliated with the Beckman Institute and Department of Physics, University of Illinois at Urbana-Champaign, Urbana, Illinois, USA. In addition, in Figure 5c, one of the structures was incorrectly labeled. Kv1.2 ch is shown in purple in the left panel of Figure 5c.The errors have been corrected for the print, PDF and HTML versions of this article.
References
Hodgkin, A.L. & Huxley, A.F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. (Lond.) 117, 500–544 (1952).
Armstrong, C.M. & Bezanilla, F. Currents related to movement of gating particles of sodium channels. Nature 242, 459–461 (1973).
Bezanilla, F. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9, 323–332 (2008).
Catterall, W.A. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron 67, 915–928 (2010).
Swartz, K.J. Sensing voltage across lipid membranes. Nature 456, 891–897 (2008).
Stühmer, W. et al. Structural parts involved in activation and inactivation of the sodium channel. Nature 339, 597–603 (1989).
Papazian, D.M., Timpe, L.C., Jan, Y.N. & Jan, L.Y. Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature 349, 305–310 (1991).
Yang, N., George, A.L. Jr. & Horn, R. Molecular basis of charge movement in voltage-gated sodium channels. Neuron 16, 113–122 (1996).
Yang, N. & Horn, R. Evidence for voltage-dependent S4 movement in sodium channels. Neuron 15, 213–218 (1995).
Mannuzzu, L.M., Moronne, M.M. & Isacoff, E.Y. Direct physical measure of conformational rearrangement underlying potassium channel gating. Science 271, 213–216 (1996).
Cha, A. & Bezanilla, F. Characterizing voltage-dependent conformational changes in the Shaker K+ channel with fluorescence. Neuron 19, 1127–1140 (1997).
Jiang, Y. et al. X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41 (2003).
Long, S.B., Campbell, E.B. & Mackinnon, R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 (2005).
Long, S.B., Tao, X., Campbell, E.B. & MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007).
Payandeh, J., Scheuer, T., Zheng, N. & Catterall, W.A. The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011).
Zhang, X. et al. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486, 130–134 (2012).
Murata, Y., Iwasaki, H., Sasaki, M., Inaba, K. & Okamura, Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435, 1239–1243 (2005).
Ramsey, I.S., Moran, M.M., Chong, J.A. & Clapham, D.E. A voltage-gated proton-selective channel lacking the pore domain. Nature 440, 1213–1216 (2006).
Sasaki, M., Takagi, M. & Okamura, Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science 312, 589–592 (2006).
Matsuda, M. et al. Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity. J. Biol. Chem. 286, 23368–23377 (2011).
Armstrong, C.M. & Bezanilla, F. Currents related to movement of the gating particles of the sodium channels. Nature 242, 459–461 (1973).
Villalba-Galea, C.A., Sandtner, W., Starace, D.M. & Bezanilla, F. S4-based voltage sensors have three major conformations. Proc. Natl. Acad. Sci. USA 105, 17600–17607 (2008).
Kohout, S.C., Ulbrich, M.H., Bell, S.C. & Isacoff, E.Y. Subunit organization and functional transitions in Ci-VSP. Nat. Struct. Mol. Biol. 15, 106–108 (2008).
Dimitrov, D. et al. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS ONE 2, e440 (2007).
Villalba-Galea, C.A., Frezza, L., Sandtner, W. & Bezanilla, F. Sensing charges of the Ciona intestinalis voltage-sensing phosphatase. J. Gen. Physiol. 142, 543–555 (2013).
Li, Q., Jogini, V., Wanderling, S., Cortes, D.M. & Perozo, E. Expression, purification, and reconstitution of the voltage-sensing domain from Ci-VSP. Biochemistry 51, 8132–8142 (2012).
Fellouse, F.A. et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 373, 924–940 (2007).
Villalba-Galea, C.A., Frezza, L., Sandtner, W. & Bezanilla, F. Sensing charges of the Ciona intestinalis voltage-sensing phosphatase. J. Gen. Physiol. 142, 543–555 (2013).
Campos, F.V., Chanda, B., Roux, B. & Bezanilla, F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc. Natl. Acad. Sci. USA 104, 7904–7909 (2007).
Tao, X., Lee, A., Limapichat, W., Dougherty, D.A. & MacKinnon, R. A gating charge transfer center in voltage sensors. Science 328, 67–73 (2010).
Schmidt, D., Jiang, Q.X. & MacKinnon, R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature 444, 775–779 (2006).
Ramu, Y., Xu, Y. & Lu, Z. Enzymatic activation of voltage-gated potassium channels. Nature 442, 696–699 (2006).
Zheng, H., Liu, W., Anderson, L.Y. & Jiang, Q.X. Lipid-dependent gating of a voltage-gated potassium channel. Nat. Commun. 2, 250 (2011).
Hack, N.J., Smith, G.P. & Peters, T.J. Subcellular localization and properties of lipase activities in human polymorphonuclear leukocytes. Biochim. Biophys. Acta 833, 406–411 (1985).
Kohout, S.C. et al. Electrochemical coupling in the voltage-dependent phosphatase Ci-VSP. Nat. Chem. Biol. 6, 369–375 (2010).
Trabuco, L.G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008).
Trabuco, L.G., Villa, E., Schreiner, E., Harrison, C.B. & Schulten, K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49, 174–180 (2009).
Ruta, V., Chen, J. & MacKinnon, R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell 123, 463–475 (2005).
Jiang, Y., Ruta, V., Chen, J., Lee, A. & MacKinnon, R. The principle of gating charge movement in a voltage-dependent K+ channel. Nature 423, 42–48 (2003).
Catterall, W.A. Molecular properties of voltage-sensitive sodium channels. Annu. Rev. Biochem. 55, 953–985 (1986).
Guy, H.R. & Seetharamulu, P. Molecular model of the action potential sodium channel. Proc. Natl. Acad. Sci. USA 83, 508–512 (1986).
Roux, B. The membrane potential and its representation by a constant electric field in computer simulations. Biophys. J. 95, 4205–4216 (2008).
Tombola, F., Pathak, M.M. & Isacoff, E.Y. How does voltage open an ion channel? Annu. Rev. Cell Dev. Biol. 22, 23–52 (2006).
Ahern, C.A. & Horn, R. Focused electric field across the voltage sensor of potassium channels. Neuron 48, 25–29 (2005).
Posson, D.J. & Selvin, P.R. Extent of voltage sensor movement during gating of shaker K+ channels. Neuron 59, 98–109 (2008).
Lin, M.C., Abramson, J. & Papazian, D.M. Transfer of ion binding site from ether-a-go-go to Shaker: Mg2+ binds to resting state to modulate channel opening. J. Gen. Physiol. 135, 415–431 (2010).
Henrion, U. et al. Tracking a complete voltage-sensor cycle with metal-ion bridges. Proc. Natl. Acad. Sci. USA 109, 8552–8557 (2012).
Vargas, E., Bezanilla, F. & Roux, B. In search of a consensus model of the resting state of a voltage-sensing domain. Neuron 72, 713–720 (2011).
Jensen, M.Ø. et al. Mechanism of voltage gating in potassium channels. Science 336, 229–233 (2012).
Arrigoni, C. et al. The voltage-sensing domain of a phosphatase gates the pore of a potassium channel. J. Gen. Physiol. 141, 389–395 (2013).
Cha, A., Snyder, G.E., Selvin, P.R. & Bezanilla, F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature 402, 809–813 (1999).
Glauner, K.S., Mannuzzu, L.M., Gandhi, C.S. & Isacoff, E.Y. Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature 402, 813–817 (1999).
Goldstein, S.A. A structural vignette common to voltage sensors and conduction pores: canaliculi. Neuron 16, 717–722 (1996).
Koag, M.C. & Papazian, D.M. Voltage-dependent conformational changes of KVAP S4 segment in bacterial membrane environment. Channels (Austin) 3, 356–365 (2009).
Li, Q., Wanderling, S., Sompornpisut, P. & Perozo, E. Structural basis of lipid-driven conformational transitions in the KvAP voltage-sensing domain. Nat. Struct. Mol. Biol. 21, 160–166 (2014).
Smart, O.S., Neduvelil, J.G., Wang, X., Wallace, B.A. & Sansom, M.S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996).
Rizk, S.S. et al. Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins. Nat. Struct. Mol. Biol. 18, 437–442 (2011).
de Vos, A.M. et al. Crystal structure of the kringle 2 domain of tissue plasminogen activator at 2.4-A resolution. Biochemistry 31, 270–279 (1992).
Finer-Moore, J.S., Kossiakoff, A.A., Hurley, J.H., Earnest, T. & Stroud, R.M. Solvent structure in crystals of trypsin determined by X-ray and neutron diffraction. Proteins 12, 203–222 (1992).
Schreiner, E., Trabuco, L.G., Freddolino, P.L. & Schulten, K. Stereochemical errors and their implications for molecular dynamics simulations. BMC Bioinformatics 12, 190 (2011).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Abelev, B.I. et al. Longitudinal double-spin asymmetry for inclusive jet production in + collisions at √s = 200 GeV. Phys. Rev. Lett. 100, 232003 (2008).
Khalili-Araghi, F. et al. Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys. J. 98, 2189–2198 (2010).
Klauda, J.B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
MacKerell, A.D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998).
Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impey, R.W. & Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Phillips, J.C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Shaw, D.E. et al. Anton, a special-purpose machine for molecular dynamics simulation. Commun. ACM 51, 91–97 (2008).
Feller, S.E. & MacKerell, A.D. An improved empirical potential energy function for molecular simulations of phospholipids. J. Phys. Chem. B 104, 7510–7515 (2000).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996).
Acknowledgements
We are thankful to K. Rajashankar and the staff at the NE-CAT 24-ID beamline as well as to R. Sanishvili and the staff at the GM/CA 23-ID beamline in the Advanced Photon Source, Argonne National Laboratory. We thank the Perozo, Bezanilla and Roux laboratories for illuminating discussions and invaluable experimental advice and A. Koide and S. Koide (University of Chicago) for the antibody library. We are grateful to E.J. Adams, R. Keenan, P. Rice and X. Yang for helpful crystallographic advice. The US National Resource provided Anton computer time for Biomedical Supercomputing and the Pittsburgh Supercomputing Center through grants RC2GM093307 and PSCA13070P (to E.P.) from the US National Institutes of Health. This work was supported in part by US National Institutes of Health grants R01-GM57846 (to E.P.), U54-GM74946 (to E.P.), R01-GM062342 (to B.R.), 9P41-GM104601 (to K.S.), U54-GM087519 (to K.S.) and 5R01-GM098243-02 (to K.S.) and a Beckman Postdoctoral Fellowship to A.S.
Author information
Authors and Affiliations
Contributions
E.P. and Q.L. designed experiments. Q.L. and S.W. performed biochemical experiments. M.P., Q.L. and A.K. selected Fab by phage-display methods. Q.L., S.W., R.E.H. and E.P. collected crystallographic data. Q.L. analyzed crystallographic data. D.M. and B.R. performed MD simulations to estimate the gating charge. A.S., R.M. and K.S. developed and applied the xMDFF method. C.A.V.-G. and Q.L. performed electrophysiological measurements. E.P. and Q.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Ci-VSD R217E structure details at 2.5-Å resolution.
(a) The Ci-VSD-R217E+33F12_4 crystal belongs to space group P6522 and contains 12 complexes (6 homodimers) in the unit cell. The dimer is formed with a crystallographically related symmetry partner on a screw axis. (b) 2Fo-Fc electron density maps (σ=1.0) of Ci-VSD-R217E complex. The S4 segment is colored in red. (c) Close-up view of R217E structure. The side chains are well resolved at 2.5 Å resolution throughout Ci-VSD including the gating arginine residues (magenta) R223, R226, R229 and R232. (d) Three LDAO detergent molecules and one succinic acid molecule were resolved around S3-S4 loop region of R217E structure (yellow sticks).
Supplementary Figure 2 EPR spectra for all scanned residues.
Continuous wave EPR spectra of spin labeled single cysteine Ci-VSD-1-260 mutants on both WT (red) and R217E (black) background scanning the region 201-250. Each pair of spectra was normalized by double integration. The largest difference in spectra shape occurred at the end of the S4-phosphatse linker which is consistent with the heterogeneity of the linker from the crystal structures.
Supplementary Figure 3 Ci-VSD WT structure details at 3.6-Å resolution.
(a) Ci-VSD WT+39D10_18 crystal belongs to space group P1 and contains 4 complexes (2 dimers) per asymmetric unit. B. Backbone of Ci-VSD WT. (b) Cα is shown in sphere for the S4 arginines (blue to cyan) hydrophobic gasket (yellow) and counter charges (red). (c) 2Fo-Fc electron density maps (σ=1.0) of one representative WT+39D10_18 complex. The S4 segment and S4-phosphatase linker were colored in red. The backbone of Ci-VSD was well resolved. (d) Superposition of the four copies of the Ci-VSDWT+39D10_18 complex inside the P1 unit cell. Backbones for the three individual domains were aligned separately and overlapped with each other within the four copies (left). Dramatic differences showed up among the four complexes when they were aligned at Ci-VSD as a whole unit (right). The variation in the four variable domains is only 1~2 Å, but extends to ~12 Å among constant domains. This apparent flexibility comes from the relative position between individual structural domains, particularly between the constant and variable domains of the Fab. The individual transmembrane regions of Ci-VSD WT are shown to be essentially identical when superimposed. There is clear heterogeneity of S4-phosphtase linker in Ci-VSD-WT since it is resolved in only three out of four copies in the asymmetric unit (e), even though the interactions involved in S4 are the same among four copies.
Supplementary Figure 4 Evaluation structural models with experimental density map.
(a) 2Fo-Fc map (at σ=1.0, teal) for residues 196-244 of Ci-VSD WT (yellow ribbon) covering the top of S3, S3-S4 loop, entire S4 and S4-phosphatase linker. The R217E structure (white ribbon) with residues 196-236 was shown for comparison. The bulky residues distributed throughout the whole region were shown as F199, Y200, E205, R223 and F234. (b) The hypothetical two-click down model (red ribbon) shift the S4 helix 3-residue downward, in reference to Ci-VSD WT model (yellow ribbon). The resulting shorter S3-S4 loop obviously deviates away from the continuous electron density map (within the red circle) and proves itself an improper model for current data. 2Fo-Fc (at σ = 1.0, teal) and Fo-Fc (at σ = 3.0, white) maps of S4+linker for Ci-VSD-WT (c) and hypothetical Up-conformation model (d). The Up-conformation model shift S4 helix 3-residue upward which leaves unaccounted electron density after residue 244 the end of current crystallization construct for Ci-VSD, by both 2Fo-Fc and Fo-Fc maps. The map quality around linker 238-244 in hypothetical “up-state” model was shown in (e) carved at 10 Å around the linker. The positive difference map (Fo-Fc) at end of the linker (white region inside red circle) is clearly above noise level.
Supplementary Figure 5 A conceptual and technical workflow of xMDFF implementation.
It shows the flexible fitting of an atomic model into an iteratively updated electron density map synthesized from experimental diffraction data, and the phase information obtained from the up-to-date fitted model. (c) Improvements in R-free, MolProbity scores and percentage of Ramachandran favored conformations resulting from xMDFF refinements of six low-resolution (4-4.5Å) X-ray structures.
Supplementary Figure 6 Rotamer orientation of gating arginines is a common motif among voltage sensors.
A large span in the relative orientation of the arginine rotamers was clearly visible if the four gating arginine residues (R223: blue, R226: lilac, R229: teal and R232: cyan) were backbone aligned in the R217E structure. R229 lies horizontally inside the hydrophobic gasket and separates the intracellular and extracellular sides electrically. R223 and R226 point straight up above the electric field, while R232 points down below the electrical field. The span of arginine rotamers was most obvious in Ci-VSD, but is also present in other existing VSDs: KvAP, Kv1.2-2.1 chimera and NavAb (however, not in NavRh). This rotameric reorientation mechanism might be an additional contributor the total gating charge translocation in VSDs.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1271 kb)
Rights and permissions
About this article
Cite this article
Li, Q., Wanderling, S., Paduch, M. et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol 21, 244–252 (2014). https://doi.org/10.1038/nsmb.2768
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2768
This article is cited by
-
Ion currents through the voltage sensor domain of distinct families of proteins
Journal of Biological Physics (2023)
-
Inhibiting Hv1 channel in peripheral sensory neurons attenuates chronic inflammatory pain and opioid side effects
Cell Research (2022)
-
Molecular dynamics simulations suggest possible activation and deactivation pathways in the hERG channel
Communications Biology (2022)
-
Complex effects on CaV2.1 channel gating caused by a CACNA1A variant associated with a severe neurodevelopmental disorder
Scientific Reports (2022)
-
Quaternary structure independent folding of voltage-gated ion channel pore domain subunits
Nature Structural & Molecular Biology (2022)