Abstract
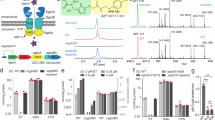
PnuC transporters catalyze cellular uptake of the NAD+ precursor nicotinamide riboside (NR) and belong to a large superfamily that includes the SWEET sugar transporters. We present a crystal structure of Neisseria mucosa PnuC, which adopts a highly symmetrical fold with 3 + 1 + 3 membrane topology not previously observed in any protein. The high symmetry of PnuC with a single NR bound in the center suggests a simple alternating-access translocation mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Kemmer, G. et al. NadN and e (P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J. Bacteriol. 183, 3974–3981 (2001).
Grose, J.H. et al. Assimilation of nicotinamide mononucleotide requires periplasmic AphA phosphatase in Salmonella enterica. J. Bacteriol. 187, 4521–4530 (2005).
Rodionov, D.A. et al. Transcriptional regulation of NAD metabolism in bacteria: genomic reconstruction of NiaR (YrxA) regulon. Nucleic Acids Res. 36, 2032–2046 (2008).
Merdanovic, M., Sauer, E. & Reidl, J. Coupling of NAD+ biosynthesis and nicotinamide ribosyl transport: characterization of NadR ribonucleotide kinase mutants of Haemophilus influenzae. J. Bacteriol. 187, 4410–4420 (2005).
Herbert, M. et al. Nicotinamide ribosyl uptake mutants in Haemophilus influenzae. Infect. Immun. 71, 5398–5401 (2003).
Gerlach, G. & Reidl, J. NAD+ utilization in Pasteurellaceae: simplification of a complex pathway. J. Bacteriol. 188, 6719–6727 (2006).
Chen, L.-Q. et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532 (2010).
Yee, D.C. et al. The transporter-opsin-G protein-coupled receptor (TOG) superfamily. FEBS J. 280, 5780–5800 (2013).
Wright, E.M. Glucose transport families SLC5 and SLC50. Mol. Aspects Med. 34, 183–196 (2013).
Lin, I.W. et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508, 546–549 (2014).
Chen, L.-Q. et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211 (2012).
Xuan, Y.H. et al. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc. Natl. Acad. Sci. USA 110, E3685–E3694 (2013).
Xu, Y. et al. Structures of bacterial homologues of SWEET transporters in two distinct conformations. Nature doi:10.1038/nature13670 (3 September 2014).
von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 7, 909–918 (2006).
Rapp, M., Granseth, E., Seppälä, S. & von Heijne, G. Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 13, 112–116 (2006).
Forrest, L.R. Structural biology: (pseudo-)symmetrical transport. Science 339, 399–401 (2013).
Sauer, E., Merdanovic, M., Mortimer, A.P., Bringmann, G. & Reidl, J. PnuC and the utilization of the nicotinamide riboside analog 3-aminopyridine in Haemophilus influenzae. Antimicrob. Agents Chemother. 48, 4532–4541 (2004).
Hemberger, S. et al. RibM from Streptomyces davawensis is a riboflavin/roseoflavin transporter and may be useful for the optimization of riboflavin production strains. BMC Biotechnol. 11, 119 (2011).
Birkner, J.P., Poolman, B. & Koçer, A. Hydrophobic gating of mechanosensitive channel of large conductance evidenced by single-subunit resolution. Proc. Natl. Acad. Sci. USA 109, 12944–12949 (2012).
Berntsson, R.P.-A. et al. Selenomethionine incorporation in proteins expressed in Lactococcus lactis. Protein Sci. 18, 1121–1127 (2009).
Ter Beek, J., Duurkens, R.H., Erkens, G.B. & Slotboom, D.J. Quaternary structure and functional unit of energy coupling factor (ECF)-type transporters. J. Biol. Chem. 286, 5471–5475 (2011).
Slotboom, D.J., Duurkens, R.H., Olieman, K. & Erkens, G.B. Static light scattering to characterize membrane proteins in detergent solution. Methods 46, 73–82 (2008).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Terwilliger, T.C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009).
Terwilliger, T.C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Murshudov, G.N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011).
Acknowledgements
We thank K. Luck for help with protein production and A.J.M. Driessen for use of his ITC machine. The research leading to these results received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (to D.J.S. and A.G., grant agreement 283570). This work was funded by the Netherlands Organisation for Scientific Research (NWO) (NWO ECHO grant 711.011.001 and NWO Vici grant 865.11.001 to D.J.S.) and the European Research Council (ERC) (ERC Starting Grant 282083 to D.J.S.). The European Synchrotron Radiation Facility (ESRF) and the Swiss Light Source (SLS) are acknowledged for beamline facilities.
Author information
Authors and Affiliations
Contributions
All authors designed experiments, performed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 2 Alignment of PnuC proteins.
The residues are colored according to their conservation (the darker the color the better the conservation). The positions of the TMs are shown as colored bars above the sequences, with the color-coding as in Figure 1. Residues involved in substrate binding are indicated by asterisks, and gate residues discussed in the text are indicated by the hash signs. The hydrophilic ladder on TM5 is indicated by a box. The numbers refer to the PnuC sequence from N. mucosa.
Supplementary Figure 3 Structural details of PnuC.
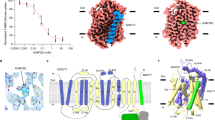
(a) Experimental anomalous map (countered at 6σ), indicating positions of 24 Selenium atoms. The backbone of the PnuC trimer is shown in gray.
(b) 2Fo-Fc electron density map (contoured at 2σ) for PnuC. Only one protomer is shown for clarity.
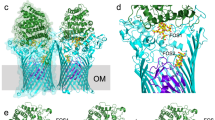
(c) Tryptophan residues and detergent molecules. The approximate position of the membrane boundaries (Fig. 1b thickness ∼27 Å) was defined by the Trp residues (shown as magenta spheres excluding Trp182 and Trp185, which are located in the binding pocket) and bound detergent molecules (n-octyl-ß-D-glucoside, cyan and red spheres). Tryptophans are preferably found in the headgroup regions of lipid bilayers.
Supplementary Figure 4 Static light scattering (SEC-MALLS) analysis of PnuC from E. coli (24 kDa, top) and Paenibacillus sp. (25 kDa, bottom).
The chromatogram from a size exclusion experiment (black line) and the calculated molar masses of the protein (red), the detergent micelle (DDM, green) and the protein-detergent micelle (blue) are shown. The experiment shows that both proteins are monomeric in detergent solution.
Supplementary Figure 5 The substrate-binding site of PnuC.
(a) Stereo image of 2Fo-Fc electron density map (contoured at 1.5σ) for the bound NR molecule and side chains of amino acids forming the binding site.
(b) Sliced-through surface representation of PnuC showing TM3 (green), TM 7 (red) and the NR molecule (yellow). On the periplasmic side two symmetry-related tyrosines (Tyr95 and Tyr217) are located at the same height as the 5'OH group of the ribose, but do not close the binding pocket. Instead, a cavity extends for one more turn of the TMs.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Table 1 (PDF 17577 kb)
Rights and permissions
About this article
Cite this article
Jaehme, M., Guskov, A. & Slotboom, D. Crystal structure of the vitamin B3 transporter PnuC, a full-length SWEET homolog. Nat Struct Mol Biol 21, 1013–1015 (2014). https://doi.org/10.1038/nsmb.2909
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2909
This article is cited by
-
The ins and outs of SWEETs in plants: Current understanding of the basics and their prospects in crop improvement
Journal of Biosciences (2021)
-
Genome-wide identification and expression analysis of the StSWEET family genes in potato (Solanum tuberosum L.)
Genes & Genomics (2020)
-
Pseudo-Symmetric Assembly of Protodomains as a Common Denominator in the Evolution of Polytopic Helical Membrane Proteins
Journal of Molecular Evolution (2020)
-
Cysteine-mediated decyanation of vitamin B12 by the predicted membrane transporter BtuM
Nature Communications (2018)
-
Solution structure and elevator mechanism of the membrane electron transporter CcdA
Nature Structural & Molecular Biology (2018)