Abstract
Processing of transcribed precursor ribosomal RNA (pre-rRNA) to a mature state is a conserved aspect of ribosome biogenesis in vivo. We developed an affinity-purification system to isolate and analyze in vivo–formed pre-rRNA–containing ribonucleoprotein (RNP) particles (rRNPs) from wild-type E. coli. We observed that the first processing intermediate of pre–small subunit (pre-SSU) rRNA is a platform for biogenesis. These pre-SSU–containing RNPs have differing ribosomal-protein and auxiliary factor association and rRNA folding. Each RNP lacks the proper architecture in functional regions, thus suggesting that checkpoints preclude immature subunits from entering the translational cycle. This work offers in vivo snapshots of SSU biogenesis and reveals that multiple pathways exist for the entire SSU biogenesis process in wild-type E. coli. These findings have implications for understanding SSU biogenesis in vivo and offer a general strategy for analysis of RNP biogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Kaczanowska, M. & Ryden-Aulin, M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 71, 477–494 (2007).
Kressler, D., Hurt, E. & Bassler, J. Driving ribosome assembly. Biochim. Biophys. Acta 1803, 673–683 (2010).
Borg, S. et al. Novel Salmonella typhimurium properties in host-parasite interactions. Immunol. Lett. 68, 247–249 (1999).
Björkman, J., Samuelsson, P., Andersson, D.I. & Hughes, D. Novel ribosomal mutations affecting translational accuracy, antibiotic resistance and virulence of Salmonella typhimurium. Mol. Microbiol. 31, 53–58 (1999).
Phunpruch, S. et al. A role for 16S rRNA dimethyltransferase (ksgA) in intrinsic clarithromycin resistance in Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 41, 548–551 (2013).
Ilina, E.N. et al. Mutation in ribosomal protein S5 leads to spectinomycin resistance in Neisseria gonorrhoeae. Front. Microbiol. 4, 186 (2013).
Champney, W.S. The other target for ribosomal antibiotics: inhibition of bacterial ribosomal subunit formation. Infect. Disord. Drug Targets 6, 377–390 (2006).
Falaleeva, M. & Stamm, S. Processing of snoRNAs as a new source of regulatory non-coding RNAs: snoRNA fragments form a new class of functional RNAs. BioEssays 35, 46–54 (2013).
Srivastava, A.K. & Schlessinger, D. Mechanism and regulation of bacterial ribosomal RNA processing. Annu. Rev. Microbiol. 44, 105–129 (1990).
Kindler, P., Keil, T.U. & Hofschneider, P.H. Isolation and characterization of a ribonuclease III deficient mutant of Escherichia coli. Mol. Gen. Genet. 126, 53–59 (1973).
Young, R.A. & Steitz, J.A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc. Natl. Acad. Sci. USA 75, 3593–3597 (1978).
Deutscher, M.P. Maturation and degradation of ribosomal RNA in bacteria. Prog. Mol. Biol. Transl. Sci. 85, 369–391 (2009).
Li, Z., Pandit, S. & Deutscher, M.P. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 18, 2878–2885 (1999).
Wachi, M., Umitsuki, G., Shimizu, M., Takada, A. & Nagai, K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 259, 483–488 (1999).
Sulthana, S. & Deutscher, M.P. Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA). J. Biol. Chem. 288, 12574–12579 (2013).
Jacob, A.I., Kohrer, C., Davies, B.W., RajBhandary, U.L. & Walker, G.C. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol. Cell 49, 427–438 (2013).
Connolly, K. & Culver, G. Deconstructing ribosome construction. Trends Biochem. Sci. 34, 256–263 (2009).
Shajani, Z., Sykes, M.T. & Williamson, J.R. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80, 501–526 (2011).
Connolly, K. & Culver, G. Overexpression of RbfA in the absence of the KsgA checkpoint results in impaired translation initiation. Mol. Microbiol. 87, 968–981 (2013).
Lindahl, L. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol. 92, 15–37 (1975).
Youngman, E.M. & Green, R. Affinity purification of in vivo–assembled ribosomes for in vitro biochemical analysis. Methods 36, 305–312 (2005).
Youngman, E.M., Brunelle, J.L., Kochaniak, A.B. & Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117, 589–599 (2004).
Persaud, C. et al. Mutagenesis of the modified bases, m(5)U1939 and psi2504, in Escherichia coli 23S rRNA. Biochem. Biophys. Res. Commun. 392, 223–227 (2010).
Asai, T. et al. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181, 3803–3809 (1999).
Moazed, D., Stern, S. & Noller, H.F. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J. Mol. Biol. 187, 399–416 (1986).
Dammel, C.S. & Noller, H.F. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 7, 660–670 (1993).
Roy-Chaudhuri, B., Kirthi, N. & Culver, G.M. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc. Natl. Acad. Sci. USA 107, 4567–4572 (2010).
Powers, T. & Noller, H.F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 10, 2203–2214 (1991).
Vila, A., Viril-Farley, J. & Tapprich, W.E. Pseudoknot in the central domain of small subunit ribosomal RNA is essential for translation. Proc. Natl. Acad. Sci. USA 91, 11148–11152 (1994).
Poot, R.A., van den Worm, S.H., Pleij, C.W. & van Duin, J. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res. 26, 549–553 (1998).
Moazed, D. & Noller, H.F. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16S rRNA. J. Mol. Biol. 211, 135–145 (1990).
Yusupova, G.Z., Yusupov, M.M., Cate, J.H. & Noller, H.F. The path of messenger RNA through the ribosome. Cell 106, 233–241 (2001).
Noller, H.F. et al. Structure of the ribosome at 5.5Å resolution and its interactions with functional ligands. Cold Spring Harb. Symp. Quant. Biol. 66, 57–66 (2001).
Schuwirth, B.S. et al. Structures of the bacterial ribosome at 3.5Å resolution. Science 310, 827–834 (2005).
Nguyenle, T., Laurberg, M., Brenowitz, M. & Noller, H.F. Following the dynamics of changes in solvent accessibility of 16S and 23S rRNA during ribosomal subunit association using synchrotron-generated hydroxyl radicals. J. Mol. Biol. 359, 1235–1248 (2006).
Culver, G.M. Assembly of the 30S ribosomal subunit. Biopolymers 68, 234–249 (2003).
Chen, S.S., Sperling, E., Silverman, J.M., Davis, J.H. & Williamson, J.R. Measuring the dynamics of E. coli ribosome biogenesis using pulse-labeling and quantitative mass spectrometry. Mol. Biosyst. 8, 3325–3334 (2012).
Mulder, A.M. et al. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science 330, 673–677 (2010).
Ogle, J.M., Carter, A.P. & Ramakrishnan, V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28, 259–266 (2003).
Kirthi, N., Roy-Chaudhuri, B., Kelley, T. & Culver, G.M. A novel single amino acid change in small subunit ribosomal protein S5 has profound effects on translational fidelity. RNA 12, 2080–2091 (2006).
Calidas, D., Lyon, H. & Culver, G.M. The N-terminal extension of S12 influences small ribosomal subunit assembly in Escherichia coli. RNA 20, 321–330 (2014).
Held, W.A., Mizushima, S. & Nomura, M. Reconstitution of Escherichia coli 30S ribosomal subunits from purified molecular components. J. Biol. Chem. 248, 5720–5730 (1973).
Chen, S.S. & Williamson, J.R. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J. Mol. Biol. 425, 767–779 (2013).
Adilakshmi, T., Bellur, D.L. & Woodson, S.A. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455, 1268–1272 (2008).
Yates, L.A., Norbury, C.J. & Gilbert, R.J. The long and short of microRNA. Cell 153, 516–519 (2013).
Mori, H., Dammel, C., Becker, E., Triman, K. & Noller, H.F. Single base alterations upstream of the E. coli 16S rRNA coding region result in temperature-sensitive 16S rRNA expression. Biochim. Biophys. Acta 1050, 323–327 (1990).
Balzer, M. & Wagner, R. Mutations in the leader region of ribosomal RNA operons cause structurally defective 30S ribosomes as revealed by in vivo structural probing. J. Mol. Biol. 276, 547–557 (1998).
Besançon, W. & Wagner, R. Characterization of transient RNA-RNA interactions important for the facilitated structure formation of bacterial ribosomal 16S RNA. Nucleic Acids Res. 27, 4353–4362 (1999).
Jomaa, A. et al. Understanding ribosome assembly: the structure of in vivo assembled immature 30S subunits revealed by cryo-electron microscopy. RNA 17, 697–709 (2011).
Leong, V., Kent, M., Jomaa, A. & Ortega, J. Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. RNA 19, 789–802 (2013).
Guo, Q. et al. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res. 41, 2609–2620 (2013).
Clatterbuck Soper, S.F., Dator, R.P., Limbach, P.A. & Woodson, S.A. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell 52, 506–516 (2013).
Bubunenko, M., Baker, T. & Court, D.L. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 189, 2844–2853 (2007).
Mizushima, S. & Nomura, M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226, 1214 (1970).
Held, W.A., Ballou, B., Mizushima, S. & Nomura, M. Assembly mapping of 30S ribosomal proteins from Escherichia coli: further studies. J. Biol. Chem. 249, 3103–3111 (1974).
Bubunenko, M. et al. 30S ribosomal subunits can be assembled in vivo without primary binding ribosomal protein S15. RNA 12, 1229–1239 (2006).
Adilakshmi, T., Ramaswamy, P. & Woodson, S.A. Protein-independent folding pathway of the 16S rRNA 5′ domain. J. Mol. Biol. 351, 508–519 (2005).
Dennis, P.P., Russell, A.G. & Moniz De Sa, M. Formation of the 5′ end pseudoknot in small subunit ribosomal RNA: involvement of U3-like sequences. RNA 3, 337–343 (1997).
Talkington, M.W., Siuzdak, G. & Williamson, J.R. An assembly landscape for the 30S ribosomal subunit. Nature 438, 628–632 (2005).
Van Duin, J. & Wijnands, R. The function of ribosomal protein S21 in protein synthesis. Eur. J. Biochem. 118, 615–619 (1981).
Douthwaite, S., Powers, T., Lee, J.Y. & Noller, H.F. Defining the structural requirements for a helix in 23S ribosomal RNA that confers erythromycin resistance. J. Mol. Biol. 209, 655–665 (1989).
Sigmund, C.D., Ettayebi, M. & Morgan, E.A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 12, 4653–4663 (1984).
Makosky, P.C. & Dahlberg, A.E. Spectinomycin resistance at site 1192 in 16S ribosomal RNA of E. coli: an analysis of three mutants. Biochimie 69, 885–889 (1987).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Sun, Q., Vila-Sanjurjo, A. & O'Connor, M. Mutations in the intersubunit bridge regions of 16S rRNA affect decoding and subunit-subunit interactions on the 70S ribosome. Nucleic Acids Res. 39, 3321–3330 (2011).
LeCuyer, K.A., Behlen, L.S. & Uhlenbeck, O.C. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry 34, 10600–10606 (1995).
Xu, Z. & Culver, G.M. Differential assembly of 16S rRNA domains during 30S subunit formation. RNA 16, 1990–2001 (2010).
Al Refaii, A. & Alix, J.H. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol. Microbiol. 71, 748–762 (2009).
Xu, Z. & Culver, G.M. Chemical probing of RNA and RNA/protein complexes. Methods Enzymol. 468, 147–165 (2009).
Stern, S., Moazed, D. & Noller, H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164, 481–489 (1988).
Das, R., Laederach, A., Pearlman, S.M., Herschlag, D. & Altman, R.B. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11, 344–354 (2005).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Eisen, M.B., Spellman, P.T., Brown, P.O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868 (1998).
Saldanha, A.J. Java Treeview-extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004).
Maki, J.A., Schnobrich, D.J. & Culver, G.M. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol. Cell 10, 129–138 (2002).
Connolly, K., Rife, J.P. & Culver, G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 70, 1062–1075 (2008).
Acknowledgements
We thank D. Ermolenko and E. Phizicky for critical discussions during the preparation of the manuscript as well as the members of Culver laboratory for helpful discussions and technical advice. We thank F. Hagen and the University of Rochester Proteomics Facility for MS analysis. We thank G. Salahura for technical assistance. We thank R. Green (Johns Hopkins University) for 86M (Spur) and MS2 coat-protein overexpression constructs and M. O'Connor (University of Missouri–Kansas City) for the MC338 strain. This work was supported by US National Institutes of Health grant GM62432 to G.M.C.
Author information
Authors and Affiliations
Contributions
N.G. and G.M.C. designed the study and experiments and wrote the manuscript; N.G. performed experiments and analyzed the data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Analysis of MS2-tag insertion on growth at low temperature and incorporation into 70S ribosomes.
(a) Schematics of 16S rRNA indicating the positions of different MS2 tags in leader, trailer and mature sequence of 16S rRNA. The tags are explained in Fig. 1b in main text, in addition, tag 86L is 86nts into the Leader and between the RNase III and RNase E cleavage sites. (b) Growth of E. coli harboring different tagged and non-tagged, wild-type (WT) plasmids at non-permissive temperature (25°C). (c) Sucrose gradient sedimentation profiles of ribosomes and ribosomal subunits from wild-type strain carrying non-tagged (WT) or 105L tagged rDNA plasmid. Allele specific primer extension analysis of rRNA (for details, see Supplementary Fig. 2) from 30S and 70S ribosomal fractions collected from sucrose gradients reveals the plasmid borne 16S rRNA in each fraction and is plotted as a histogram (n = 3, standard deviation is shown as error bars).
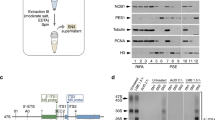
Supplementary Figure 2 Outline for the purification of tagged SSU assembly intermediates from wild-type E. coli.
MS2CP = MS2 coat protein fused with Maltose Binding Protein (MBP). Plasmid borne 16S rRNA carry the spectinomycin resistance mutation, C1192U, which is absent in genomic operons. Allele specific primer extension of 16S rRNA purified with tags at position 105L, 11L and 20T using Cy5 labeled primer starting reverse transcription at position 1193 in the presence of ddGTP and differential stops are detected on 20% polyacrylamide gel. 16S represents mature 16S rRNA from genomic operons. Quantification shows more than 95% rRNA is plasmid borne. 1192C is chromosomally encoded while 1192U is plasmid encoded 16S rRNA.
Supplementary Figure 3 Examination of nucleotides with altered reactivity in three SSU intermediates.
Nucleotides of mature 16S rRNA with altered reactivity in the three pre-SSU complexes purified with different tags are shown. The relative changes in the nucleotide reactivity intensity of the intermediates to mature subunits are plotted on a log2 scale (see Supplementary Data Set1). The dotted lines indicate cutoff values for relative change in nucleotide reactivity to be considered. The regions involved in pseudoknot formation, intersubunit bridges and tRNA interactions are marked.
Supplementary Figure 4 R-protein content of the three purified SSU intermediates is distinct.
(a, b) Interface side representation of SSU for Figs. 4a and 4b respectively in the main text. (c) The three groups emerged from the hierarchical clustering analysis are plotted on the in vitro assembly map. The dotted lines divide the three groups. The circles around the r-proteins indicate the free precursor pool of that protein in the wild-type cells as determined previously37.
Supplementary Figure 5 Delayed processing of 17S rRNA in the absence of RNase G and RNase E.
Modified 3′5′ RACE products obtained during the processing of 17S rRNA associated with 11L and 20T assembly intermediates when incubated with different cell extract in vitro are separated on agarose gel. Different S100 extracts prepared from Δrng strain or a strain harboring a ts allele of rne13 are compared to wild-type S100 extract. Incubation time (in mins) is shown.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 (PDF 2881 kb)
Supplementary Data Set 1
Chemical modification of 17S rRNA–containing RNPs using kethoxal (XLSX 50 kb)
Supplementary Data Set 2
Relative levels of R proteins in SSU intermediates (XLSX 15 kb)
Supplementary Data Set 3
Uncropped gels and northern blots shown in the main figure (PDF 28616 kb)
Rights and permissions
About this article
Cite this article
Gupta, N., Culver, G. Multiple in vivo pathways for Escherichia coli small ribosomal subunit assembly occur on one pre-rRNA. Nat Struct Mol Biol 21, 937–943 (2014). https://doi.org/10.1038/nsmb.2887
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2887
This article is cited by
-
Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis
Nature Structural & Molecular Biology (2015)