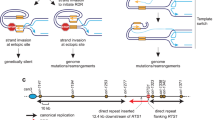
Abstract
Although homologous recombination is considered an accurate form of DNA repair, genetics suggest that the Escherichia coli translesion DNA polymerase IV (Pol IV, also known as DinB) promotes error-prone recombination during stress, which allows cells to overcome adverse conditions. However, how Pol IV functions and is regulated during recombination under stress is unknown. We show that Pol IV is highly proficient in error-prone recombination and is preferentially recruited to displacement loops (D loops) at stress-induced concentrations in vitro. We also found that high-fidelity Pol II switches to exonuclease mode at D loops, which is stimulated by topological stress and reduced deoxyribonucleotide pool concentration during stationary phase. The exonuclease activity of Pol II enables it to compete with Pol IV, which probably suppresses error-prone recombination. These findings indicate that preferential D-loop extension by Pol IV facilitates error-prone recombination and explain how Pol II reduces such errors in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Cox, M.M. Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 35, 53–82 (2001).
Cox, M.M. et al. The importance of repairing stalled replication forks. Nature 404, 37–41 (2000).
Mazon, G., Mimitou, E.P. & Symington, L.S. SnapShot: homologous recombination in DNA double-strand break repair. Cell 142, 646, 646.e1 (2010).
Li, X. & Heyer, W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113 (2008).
Heller, R.C. & Marians, K.J. Replisome assembly and the direct restart of stalled replication forks. Nat. Rev. Mol. Cell Biol. 7, 932–943 (2006).
Zahradka, K. et al. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443, 569–573 (2006).
Motamedi, M.R., Szigety, S.K. & Rosenberg, S.M. Double-strand–break repair recombination in Escherichia coli: physical evidence for a DNA replication mechanism in vivo. Genes Dev. 13, 2889–2903 (1999).
Lydeard, J.R. et al. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 24, 1133–1144 (2010).
Kawamoto, T. et al. Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol. Cell 20, 793–799 (2005).
Kane, D.P., Shusterman, M., Rong, Y. & McVey, M. Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila. PLoS Genet. 8, e1002659 (2012).
Li, X., Stith, C.M., Burgers, P.M. & Heyer, W.D. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol. Cell 36, 704–713 (2009).
Hashimoto, Y., Puddu, F. & Costanzo, V. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat. Struct. Mol. Biol. 19, 17–24 (2012).
Moldovan, G.L. et al. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol. Cell Biol. 30, 1088–1096 (2010).
McIlwraith, M.J. et al. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell 20, 783–792 (2005).
Kohzaki, M. et al. DNA polymerases ν and θ are required for efficient immunoglobulin V gene diversification in chicken. J. Cell Biol. 189, 1117–1127 (2010).
Rosenberg, S.M. Evolving responsively: adaptive mutation. Nat. Rev. Genet. 2, 504–515 (2001).
Ponder, R.G., Fonville, N.C. & Rosenberg, S.M. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol. Cell 19, 791–804 (2005).
McKenzie, G.J., Lee, P.L., Lombardo, M.J., Hastings, P.J. & Rosenberg, S.M. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7, 571–579 (2001).
Foster, P.L. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42, 373–397 (2007).
Hastings, P.J. et al. Competition of Escherichia coli DNA polymerases I, II and III with DNA Pol IV in stressed cells. PLoS ONE 5, e10862 (2010).
Shee, C., Gibson, J.L. & Rosenberg, S.M. Two mechanisms produce mutation hotspots at DNA breaks in Escherichia coli. Cell Rep. 2, 714–721 (2012).
Harris, R.S., Bull, H.J. & Rosenberg, S.M. A direct role for DNA polymerase III in adaptive reversion of a frameshift mutation in Escherichia coli. Mutat. Res. 375, 19–24 (1997).
Schlacher, K., Cox, M.M., Woodgate, R. & Goodman, M.F. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442, 883–887 (2006).
Petrosino, J.F., Galhardo, R.S., Morales, L.D. & Rosenberg, S.M. Stress-induced β-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J. Bacteriol. 191, 5881–5889 (2009).
Kim, S.R., Matsui, K., Yamada, M., Gruz, P. & Nohmi, T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol. Genet. Genomics 266, 207–215 (2001).
Tippin, B., Pham, P. & Goodman, M.F. Error-prone replication for better or worse. Trends Microbiol. 12, 288–295 (2004).
Akerlund, T., Nordstrom, K. & Bernander, R. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J. Bacteriol. 177, 6791–6797 (1995).
Withers, H.L. & Bernander, R. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J. Bacteriol. 180, 1624–1631 (1998).
Godoy, V.G. et al. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol. Cell 28, 1058–1070 (2007).
Kobayashi, S., Valentine, M.R., Pham, P., O'Donnell, M. & Goodman, M.F. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J. Biol. Chem. 277, 34198–34207 (2002).
Galhardo, R.S. et al. DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics 182, 55–68 (2009).
Xu, L. & Marians, K.J. PriA mediates DNA replication pathway choice at recombination intermediates. Mol. Cell 11, 817–826 (2003).
Shlomai, J. & Kornberg, A. A prepriming DNA replication enzyme of Escherichia coli. I. Purification of protein n′: a sequence-specific, DNA-dependent ATPase. J. Biol. Chem. 255, 6789–6793 (1980).
Frisch, R.L. et al. Separate DNA Pol II– and Pol IV–dependent pathways of stress-induced mutation during double-strand–break repair in Escherichia coli are controlled by RpoS. J. Bacteriol. 192, 4694–4700 (2010).
Foster, P.L., Gudmundsson, G., Trimarchi, J.M., Cai, H. & Goodman, M.F. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc. Natl. Acad. Sci. USA 92, 7951–7955 (1995).
Bonner, C.A. et al. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J. Biol. Chem. 263, 18946–18952 (1988).
Cai, H., Yu, H., McEntee, K., Kunkel, T.A. & Goodman, M.F. Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from Escherichia coli. J. Biol. Chem. 270, 15327–15335 (1995).
Goodman, M.F. et al. DNA Replication and Mutagenesis (American Society for Microbiology, Washington, D.C., 1988).
Buckstein, M.H., He, J. & Rubin, H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726 (2008).
Ibarra, B. et al. Proofreading dynamics of a processive DNA polymerase. EMBO J. 28, 2794–2802 (2009).
Downey, C.D. & McHenry, C.S. Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader. Mol. Cell 37, 481–491 (2010).
Baker, T. & Kornberg, A. DNA Replication, 2nd edn (University Science Books, 1992).
Lovett, S.T. Replication arrest-stimulated recombination: dependence on the RecA paralog, RadA/Sms and translesion polymerase, DinB. DNA Repair (Amst.) 5, 1421–1427 (2006).
Register, J.C. III & Griffith, J. Direct visualization of RecA protein binding to and unwinding duplex DNA following the D-loop cycle. J. Biol. Chem. 263, 11029–11032 (1988).
Indiani, C., McInerney, P., Georgescu, R., Goodman, M.F. & O'Donnell, M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell 19, 805–815 (2005).
Georgescu, R.E. et al. Mechanism of polymerase collision release from sliding clamps on the lagging strand. EMBO J. 28, 2981–2991 (2009).
Davey, M.J., Fang, L., McInerney, P., Georgescu, R.E. & O'Donnell, M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 21, 3148–3159 (2002).
Indiani, C., Langston, L.D., Yurieva, O., Goodman, M.F. & O'Donnell, M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc. Natl. Acad. Sci. USA 106, 6031–6038 (2009).
Kaplan, D.L. & O'Donnell, M. RuvA is a sliding collar that protects Holliday junctions from unwinding while promoting branch migration. J. Mol. Biol. 355, 473–490 (2006).
McInerney, P. & O'Donnell, M. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 279, 21543–21551 (2004).
Pomerantz, R.T. & O'Donnell, M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456, 762–766 (2008).
Acknowledgements
We thank M. Skangalis, D. Zhang and O. Yurieva for technical support. This work was supported by the US National Institutes of Health (grants to M.E.O. (GM38839), R.T.P. (K99CA160648) and M.F.G. (GM21422 and ES012259)) and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
R.T.P. conceived the idea for the study, wrote the manuscript and performed and interpreted all experiments, with the exception of those in Figure 3i,j, which were performed by I.K. M.E.O. and M.F.G. provided editorial input.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 870 kb)
Rights and permissions
About this article
Cite this article
Pomerantz, R., Kurth, I., Goodman, M. et al. Preferential D-loop extension by a translesion DNA polymerase underlies error-prone recombination. Nat Struct Mol Biol 20, 748–755 (2013). https://doi.org/10.1038/nsmb.2573
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2573
This article is cited by
-
Specialised DNA polymerases in Escherichia coli: roles within multiple pathways
Current Genetics (2018)
-
Collision with duplex DNA renders Escherichia coli DNA polymerase III holoenzyme susceptible to DNA polymerase IV-mediated polymerase switching on the sliding clamp
Scientific Reports (2017)
-
Single-molecule imaging reveals multiple pathways for the recruitment of translesion polymerases after DNA damage
Nature Communications (2017)