Abstract
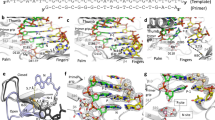
Hundreds of structures of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase (RT) have been determined, but only one contains an RNA/DNA hybrid. Here we report three structures of HIV-1 RT complexed with a non-nucleotide RT inhibitor (NNRTI) and an RNA/DNA hybrid. In the presence of an NNRTI, the RNA/DNA structure differs from all prior nucleic acid–RT structures including the RNA/DNA hybrid. The enzyme structure also differs from all previous RT–DNA complexes. Thus, the hybrid has ready access to the RNase-H active site. These observations indicate that an RT–nucleic acid complex may adopt two structural states, one competent for DNA polymerization and the other for RNA degradation. RT mutations that confer drug resistance but are distant from the inhibitor-binding sites often map to the unique RT-hybrid interface that undergoes conformational changes between two catalytic states.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Varmus, H. Reverse transcription. Sci. Am. 257, 56–59, 62–64 (1987).
Sarafianos, S.G. et al. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385, 693–713 (2009).
Cihlar, T. & Ray, A.S. Nucleoside and nucleotide HIV reverse transcriptase inhibitors: 25 years after zidovudine. Antiviral Res. 85, 39–58 (2010).
de Béthune, M.P. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989–2009). Antiviral Res. 85, 75–90 (2010).
di Marzo Veronese, F. et al. Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science 231, 1289–1291 (1986).
Lightfoote, M.M. et al. Structural characterization of reverse transcriptase and endonuclease polypeptides of the acquired immunodeficiency syndrome retrovirus. J. Virol. 60, 771–775 (1986).
Gilboa, E., Mitra, S.W., Goff, S. & Baltimore, D. A detailed model of reverse transcription and tests of crucial aspects. Cell 18, 93–100 (1979).
Whitcomb, J.M. & Hughes, S.H. Retroviral reverse transcription and integration: progress and problems. Annu. Rev. Cell Biol. 8, 275–306 (1992).
Nowotny, M., Gaidamakov, S.A., Crouch, R.J. & Yang, W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121, 1005–1016 (2005).
Nowotny, M. et al. Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol. Cell 28, 264–276 (2007).
Schultz, S.J. & Champoux, J.J. RNase H activity: structure, specificity, and function in reverse transcription. Virus Res. 134, 86–103 (2008).
Shaw-Reid, C.A. et al. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem. 278, 2777–2780 (2003).
Himmel, D.M. et al. Structure of HIV-1 reverse transcriptase with the inhibitor β-Thujaplicinol bound at the RNase H active site. Structure 17, 1625–1635 (2009).
Lansdon, E.B. et al. Structural and binding analysis of pyrimidinol carboxylic acid and N-hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob. Agents Chemother. 55, 2905–2915 (2011).
Steitz, T.A. & Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. USA 90, 6498–6502 (1993).
Yang, W., Lee, J.Y. & Nowotny, M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell 22, 5–13 (2006).
Huber, H.E. & Richardson, C.C. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J. Biol. Chem. 265, 10565–10573 (1990).
Fuentes, G.M., Rodriguez-Rodriguez, L., Fay, P.J. & Bambara, R.A. Use of an oligoribonucleotide containing the polypurine tract sequence as a primer by HIV reverse transcriptase. J. Biol. Chem. 270, 28169–28176 (1995).
Gopalakrishnan, V., Peliska, J.A. & Benkovic, S.J. Human immunodeficiency virus type 1 reverse transcriptase: spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. USA 89, 10763–10767 (1992).
Kohlstaedt, L.A., Wang, J., Friedman, J.M., Rice, P.A. & Steitz, T.A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256, 1783–1790 (1992).
Hopkins, A.L. et al. Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors. J. Med. Chem. 39, 1589–1600 (1996).
Ren, J. et al. Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase. Structure 8, 1089–1094 (2000).
Ren, J. & Stammers, D.K. Structural basis for drug resistance mechanisms for non-nucleoside inhibitors of HIV reverse transcriptase. Virus Res. 134, 157–170 (2008).
Tu, X. et al. Structural basis of HIV-1 resistance to AZT by excision. Nat. Struct. Mol. Biol. 17, 1202–1209 (2010).
Jacobo-Molina, A. et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90, 6320–6324 (1993).
Huang, H., Chopra, R., Verdine, G.L. & Harrison, S.C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282, 1669–1675 (1998).
Lansdon, E.B. et al. Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex. J. Mol. Biol. 397, 967–978 (2010).
Meyer, P.R., Matsuura, S.E., Mian, A.M., So, A.G. & Scott, W.A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4, 35–43 (1999).
Das, K., Martinez, S.E., Bauman, J.D. & Arnold, E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat. Struct. Mol. Biol. 19, 253–259 (2012).
Sarafianos, S.G. et al. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 20, 1449–1461 (2001).
Beilhartz, G.L. et al. HIV-1 reverse transcriptase can simultaneously engage its DNA/RNA substrate at both DNA polymerase and RNase H active sites: implications for RNase H inhibition. J. Mol. Biol. 388, 462–474 (2009).
Hang, J.Q. et al. Substrate-dependent inhibition or stimulation of HIV RNase H activity by non-nucleoside reverse transcriptase inhibitors (NNRTIs). Biochem. Biophys. Res. Commun. 352, 341–350 (2007).
Radzio, J. & Sluis-Cremer, N. Efavirenz accelerates HIV-1 reverse transcriptase ribonuclease H cleavage, leading to diminished zidovudine excision. Mol. Pharmacol. 73, 601–606 (2008).
Shaw-Reid, C.A. et al. Dissecting the effects of DNA polymerase and ribonuclease H inhibitor combinations on HIV-1 reverse-transcriptase activities. Biochemistry 44, 1595–1606 (2005).
Delviks-Frankenberry, K.A. et al. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc. Natl. Acad. Sci. USA 105, 10943–10948 (2008).
Delviks-Frankenberry, K.A., Nikolenko, G.N. & Pathak, V.K. The “connection” between HIV drug resistance and RNase H. Viruses 2, 1476–1503 (2010).
Peliska, J.A. & Benkovic, S.J. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258, 1112–1118 (1992).
Ding, J. et al. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 284, 1095–1111 (1998).
Hsiou, Y. et al. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure 4, 853–860 (1996).
Rodgers, D.W. et al. The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 92, 1222–1226 (1995).
Brehm, J.H., Mellors, J.W. & Sluis-Cremer, N. Mechanism by which a glutamine to leucine substitution at residue 509 in the ribonuclease H domain of HIV-1 reverse transcriptase confers zidovudine resistance. Biochemistry 47, 14020–14027 (2008).
Tachedjian, G., Orlova, M., Sarafianos, S.G., Arnold, E. & Goff, S.P. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. USA 98, 7188–7193 (2001).
Yap, S.H. et al. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 4, e335 (2007).
Ehteshami, M. et al. Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3′-azido-3′-deoxythymidine through both RNase H-dependent and -independent mechanisms. J. Biol. Chem. 283, 22222–22232 (2008).
Cameron, C.E., Ghosh, M., Le Grice, S.F. & Benkovic, S.J. Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc. Natl. Acad. Sci. USA 94, 6700–6705 (1997).
Chung, S., Miller, J.T., Johnson, B.C., Hughes, S.H. & Le Grice, S.F. Mutagenesis of human immunodeficiency virus reverse transcriptase p51 subunit defines residues contributing to vinylogous urea inhibition of ribonuclease H activity. J. Biol. Chem. 287, 4066–4075 (2012).
Perry, J.J., Cotner-Gohara, E., Ellenberger, T. & Tainer, J.A. Structural dynamics in DNA damage signaling and repair. Curr. Opin. Struct. Biol. 20, 283–294 (2010).
Rambo, R.P. & Tainer, J.A. Bridging the solution divide: comprehensive structural analyses of dynamic RNA, DNA, and protein assemblies by small-angle X-ray scattering. Curr. Opin. Struct. Biol. 20, 128–137 (2010).
Gerondelis, P. et al. The P236L delavirdine-resistant human immunodeficiency virus type 1 mutant is replication defective and demonstrates alterations in both RNA 5′-end– and DNA 3′-end–directed RNase H activities. J. Virol. 73, 5803–5813 (1999).
Wendeler, M. et al. Vinylogous ureas as a novel class of inhibitors of reverse transcriptase–associated ribonuclease H activity. ACS Chem. Biol. 3, 635–644 (2008).
Chung, S. et al. Structure-activity analysis of vinylogous urea inhibitors of human immunodeficiency virus-encoded ribonuclease H. Antimicrob. Agents Chemother. 54, 3913–3921 (2010).
Tuske, S. et al. Structures of HIV-1 RT–DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Nat. Struct. Mol. Biol. 11, 469–474 (2004).
Le Grice, S.F., Cameron, C.E. & Benkovic, S.J. Purification and characterization of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 262, 130–144 (1995).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Cohen, G.H. ALIGN: a program to superimpose protein coordinates, accounting for insertions and deletions. J. Appl. Crystallogr. 30, 1160–1161 (1997).
Richards, F.M. & Kundrot, C.E. Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. Proteins 3, 71–84 (1988).
Lu, X.J. & Olson, W.K. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 3, 1213–1227 (2008).
Lavery, R., Moakher, M., Maddocks, J.H., Petkeviciute, D. & Zakrzewska, K. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res. 37, 5917–5929 (2009).
Acknowledgements
We thank D. Leahy and M. Nowotny for editing and critiquing the manuscript, C. Biertümpfel and Y. Zhao for help with data collection and J.H.D. Cate (University of California, Berkeley, Berkeley, California, USA) for the ModeVector script to make Figures 2 and 3a. The research was supported by the US National Institutes of Health Intramural AIDS Targeted Anti-viral Program (IATAP) and the intramural research programs of the US National Institute of Diabetes and Digestive and Kidney Diseases (W.Y., M.L. and L.T.) and the US National Cancer Institute (S.F.J.L.G. and J.T.M.).
Author information
Authors and Affiliations
Contributions
J.T.M. prepared the HIV-1 RT mutants and proteins. L.T. made the RT–RNA/DNA–NNRTI complexes, grew the crystals and collected the diffraction data. M.L. refined the structures, and W.Y. carried out structure comparisons. W.Y. and S.F.J.L.G. originated the project. M.L., L.T., S.F.J.L.G. and W.Y. prepared the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 2802 kb)
Supplementary Video 1
Video 1 Structural differences between RT-NNRTI and the RT-RNA/DNA-NNRTI complex (Video1.mov) (MOV 2259 kb)
Supplementary Video 2
Video 2 Structural differences between RT-DNA-dATP and the RT-RNA/DNA-NNRTI complex. (Video2.mov) (MOV 2231 kb)
Supplementary Video 3
Video 3 Morphing of nucleic acid complexed with HIV-1 RT from the DNA polymerization to RNA degradation-compatible state. (video3.mov) (MOV 1962 kb)
Rights and permissions
About this article
Cite this article
Lapkouski, M., Tian, L., Miller, J. et al. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nat Struct Mol Biol 20, 230–236 (2013). https://doi.org/10.1038/nsmb.2485
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2485
This article is cited by
-
Transcriptional inaccuracy threshold attenuates differences in RNA-dependent DNA synthesis fidelity between retroviral reverse transcriptases
Scientific Reports (2018)
-
NMR structure of the HIV-1 reverse transcriptase thumb subdomain
Journal of Biomolecular NMR (2016)
-
Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry
Nature Structural & Molecular Biology (2014)
-
Novel insights from structural analysis of lentiviral and gammaretroviral reverse transcriptases in complex with RNA/DNA hybrids
Retrovirology (2013)
-
Structural requirements for RNA degradation by HIV-1 reverse transcriptase
Nature Structural & Molecular Biology (2013)