Abstract
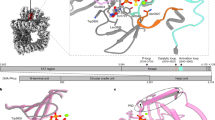
Cytoplasmic terminal uridylyl transferases comprise a conserved family of enzymes that negatively regulate the stability or biological activity of a variety of eukaryotic RNAs, including mRNAs and tumor-suppressor let-7 microRNAs. Here we describe crystal structures of the Schizosaccharomyces pombe cytoplasmic terminal uridylyl transferase Cid1 in two apo conformers and bound to UTP. We demonstrate that a single histidine residue, conserved in mammalian Cid1 orthologs, is responsible for discrimination between UTP and ATP. We also describe a new high-affinity RNA substrate–binding mechanism of Cid1, which is essential for enzymatic activity and is mediated by three basic patches across the surface of the enzyme. Overall, our structures provide a basis for understanding the activity of Cid1 and a mechanism of UTP selectivity conserved in its human orthologs, suggesting potential implications for anticancer drug design.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hagan, J.P., Piskounova, E. & Gregory, R.I. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 16, 1021–1025 (2009).
Heo, I. et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708 (2009).
Jones, M.R. et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 11, 1157–1163 (2009).
Lehrbach, N.J. et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16, 1016–1020 (2009).
Mullen, T.E. & Marzluff, W.F. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22, 50–65 (2008).
Schmidt, M.-J., West, S. & Norbury, C. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17, 39–44 (2011).
Shen, B. & Goodman, H.M. Uridine addition after microRNA-directed cleavage. Science 306, 997 (2004).
Rissland, O.S., Mikulasova, A. & Norbury, C.J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 27, 3612–3624 (2007).
Rissland, O.S. & Norbury, C.J. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16, 616–623 (2009).
Bard, J. et al. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science 289, 1346–1349 (2000).
Martin, G., Möglich, A., Keller, W. & Doublié, S. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J. Mol. Biol. 341, 911–925 (2004).
Piskounova, E. et al. Lin28A and Lin28B inhibit let-7 MicroRNA biogenesis by distinct mechanisms. Cell 147, 1066–1079 (2011).
Sawaya, M.R., Pelletier, H., Kumar, A., Wilson, S.H. & Kraut, J. Crystal structure of rat DNA polymerase β: evidence for a common polymerase mechanism. Science 264, 1930–1935 (1994).
Stagno, J., Aphasizheva, I., Rosengarth, A., Luecke, H. & Aphasizhev, R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 366, 882–899 (2007).
Riffel, N. et al. Atomic resolution structure of Moloney murine leukemia virus matrix protein and its relationship to other retroviral matrix proteins. Structure 10, 1627–1636 (2002).
Stuart, D.I., Levine, M., Muirhead, H. & Stammers, D.K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J. Mol. Biol. 134, 109–142 (1979).
Stagno, J., Aphasizheva, I., Aphasizhev, R. & Luecke, H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc. Natl. Acad. Sci. USA 104, 14634–14639 (2007).
Martin, G. & Keller, W. RNA-specific ribonucleotidyl transferases. RNA 13, 1834–1849 (2007).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Deng, J., Ernst, N.L., Turley, S., Stuart, K.D. & Hol, W.G. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 24, 4007–4017 (2005).
Hamill, S., Wolin, S.L. & Reinisch, K.M. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc. Natl. Acad. Sci. USA 107, 15045–15050 (2010).
LaCava, J. et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121, 713–724 (2005).
Vaňáčová, S. et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3, e189 (2005).
Read, R.L., Martinho, R.G., Wang, S.W., Carr, A.M. & Norbury, C.J. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. USA 99, 12079–12084 (2002).
Deo, R.C., Bonanno, J.B., Sonenberg, N. & Burley, S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98, 835–845 (1999).
Bai, Y., Srivastava, S.K., Chang, J.H., Manley, J.L. & Tong, L. Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase. Mol. Cell 41, 311–320 (2011).
Hayward, S. & Berendsen, H.J. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30, 144–154 (1998).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010).
Leslie, A.G.W. & Powell, H.R. in Evolving Methods for Macromolecular Crystallography, Nato Science Series II Vol. 245 (eds Read, R.J. & Sussman, J.L.) 41–51 (Springer, 2007).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Stein, N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Crystallogr. 41, 641–643 (2008).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Zhang, K.Y., Cowtan, K. & Main, P. Combining constraints for electron-density modification. Methods Enzymol. 277, 53–64 (1997).
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Painter, J. & Merritt, E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 (2006).
Kleywegt, G.J. Crystallographic refinement of ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 63, 94–100 (2007).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Pettersen, E.F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
The authors thank N. Proudfoot, A. van der Merwe, F. Esashi and T. Schiffner for discussions and comments on the manuscript, A. Watson and J. Endicott for their input in the early stages of this project, and Cancer Research UK (C.J.N.), The Royal Society (R.J.C.G.), the Wellcome Trust (core-Centre grant 090532/Z/09/Z), the Medical Research Council (L.A.Y. and K.H.) and the EP Abraham Research Fund (C.J.N.) for financial support.
Author information
Authors and Affiliations
Contributions
The project was initiated by O.S.R., L.A.Y., C.J.N. and R.J.C.G.; C.J.N and R.J.C.G. designed and supervised the project. O.S.R. designed the tCid1 expression construct. S.F. characterized the RNA-binding capacity of Cid1, designed and purified the tCid1 mutants and designed and performed EMSA, TUTase assays and SPR. L.A.Y. purified, crystallized tCid1 and optimized crystals for data collection. K.H. soaked and handled crystals for data collection. L.A.Y. collected, processed and analyzed the X-ray diffraction data. L.D.C. solved the initial structure; L.A.Y. solved subsequent structures and built and refined all tCid1 structures. L.A.Y. and R.J.C.G. performed the structural and phylogenetic analysis. L.A.Y., S.F., C.J.N. and R.J.C.G. wrote the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5, Supplementary Tables 1–4 and Supplementary Note (PDF 4937 kb)
Supplementary Video 1
Morphing between the Apo I, UTP-bound and Apo II structures (salmon pink) superimposed on the UTP-bound structure (yellow, UTP in green), viewed into active site cleft. (AVI 2118 kb)
Supplementary Video 2
As Supplementary Video 1, viewed from side. (AVI 1924 kb)
Rights and permissions
About this article
Cite this article
Yates, L., Fleurdépine, S., Rissland, O. et al. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat Struct Mol Biol 19, 782–787 (2012). https://doi.org/10.1038/nsmb.2329
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2329
This article is cited by
-
Mechanism of U6 snRNA oligouridylation by human TUT1
Nature Communications (2023)
-
A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing
Nature Reviews Molecular Cell Biology (2020)
-
Crystal structure of the Lin28-interacting module of human terminal uridylyltransferase that regulates let-7 expression
Nature Communications (2019)
-
Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase
Nature Methods (2019)
-
Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nature Communications (2017)