Abstract
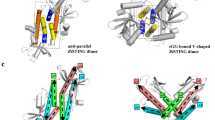
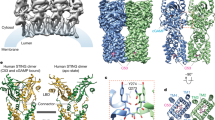
STING functions as both an adaptor protein signaling cytoplasmic double-stranded DNA and a direct immunosensor of cyclic diguanylate monophosphate (c-di-GMP). The crystal structures of the C-terminal domain of human STING (STINGCTD) and its complex with c-di-GMP reveal how STING recognizes c-di-GMP. In response to c-di-GMP binding, two surface loops, which serve as a gate and latch of the cleft formed by the dimeric STINGCTD, undergo rearrangements to interact with the ligand.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kumar, H., Kawai, T. & Akira, S. Int. Rev. Immunol. 30, 16–34 (2011).
Kawasaki, T., Kawai, T. & Akira, S. Immunol. Rev. 243, 61–73 (2011).
Hornung, V. & Latz, E. Nat. Rev. Immunol. 10, 123–130 (2010).
Ishikawa, H. & Barber, G.N. Nature 455, 674–678 (2008).
Ishikawa, H., Ma, Z. & Barber, G.N. Nature 461, 788–792 (2009).
Jin, L. et al. Mol. Cell Biol. 28, 5014–5026 (2008).
Zhong, B. et al. Immunity 29, 538–550 (2008).
Sun, W. et al. Proc. Natl. Acad. Sci. USA 106, 8653–8658 (2009).
Tamayo, R., Pratt, J.T. & Camilli, A. Annu. Rev. Microbiol. 61, 131–148 (2007).
Karaolis, D.K. et al. J. Immunol. 178, 2171–2181 (2007).
McWhirter, S.M. et al. J. Exp. Med. 206, 1899–1911 (2009).
Sauer, J.D. et al. Infect. Immun. 79, 688–694 (2011).
Woodward, J.J., Iavarone, A.T. & Portnoy, D.A. Science 328, 1703–1705 (2010).
Jin, L. et al. J. Immunol. 187, 2595–2601 (2011).
Abdul-Sater, A.A. et al. Microbes Infect. 14, 188–197 (2012).
Burdette, D.L. et al. Nature 478, 515–518 (2011).
Holm, L. & Rosenstrom, P. Nucleic Acids Res. 38, W545–W549 (2010).
Benach, J. et al. EMBO J. 26, 5153–5166 (2007).
Jin, L. et al. Genes Immun. 12, 263–269 (2011).
Tsuchida, T. et al. Immunity 33, 765–776 (2010).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project Number 4. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Adams, P.D. et al. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Terwilliger, T.C. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 (2003).
Emsley, P. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
McCoy, A.J. et al. J. Appl. Crystallogr. 40, 658–674 (2007).
Chenna, R. et al. Nucleic Acids Res. 31, 3497–3500 (2003).
Gouet, P., Courcelle, E., Stuart, D.I. & Metoz, F. Bioinformatics 15, 305–308 (1999).
Acknowledgements
We thank R. Qiu for providing human cDNA library, the staff at Shanghai Synchrotron Radiation Facility beamline 17U for assistance with data collection. This work was supported by the State Key Laboratory of Microbial Technology, Shandong University, by Hi-Tech Research and Development Program of China grant no. 2006AA02A324 to L.G. and by National Nature Science Foundation of China grant no. 31000330 to D.Z.
Author information
Authors and Affiliations
Contributions
G.S., D.Z., N.L., J.Z. and L.G. designed the research; G.S., D.Z., N.L., J.Z., C.Z., D.L., C.L., Q.Y. and Y.Z. performed the experiments; G.S., D.Z., N.L., J.Z., S.X. and L.G. analyzed data and wrote the paper; all authors contributed to the editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Table 1 (PDF 1564 kb)
Rights and permissions
About this article
Cite this article
Shang, G., Zhu, D., Li, N. et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol 19, 725–727 (2012). https://doi.org/10.1038/nsmb.2332
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2332
This article is cited by
-
cGAMP-activated cGAS–STING signaling: its bacterial origins and evolutionary adaptation by metazoans
Nature Structural & Molecular Biology (2023)
-
Molecular mechanisms of mitochondrial DNA release and activation of the cGAS-STING pathway
Experimental & Molecular Medicine (2023)
-
Crystal structure and functional implication of bacterial STING
Nature Communications (2022)
-
Molecular mechanisms and cellular functions of cGAS–STING signalling
Nature Reviews Molecular Cell Biology (2020)
-
STING cyclic dinucleotide sensing originated in bacteria
Nature (2020)