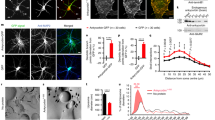
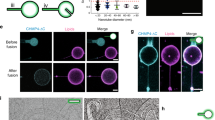
Abstract
Bin/amphipysin/Rvs (BAR)-domain proteins sculpt cellular membranes and have key roles in processes such as endocytosis, cell motility and morphogenesis. BAR domains are divided into three subfamilies: BAR– and F-BAR–domain proteins generate positive membrane curvature and stabilize cellular invaginations, whereas I-BAR–domain proteins induce negative curvature and stabilize protrusions. We show that a previously uncharacterized member of the I-BAR subfamily, Pinkbar, is specifically expressed in intestinal epithelial cells, where it localizes to Rab13-positive vesicles and to the plasma membrane at intercellular junctions. Notably, the BAR domain of Pinkbar does not induce membrane tubulation but promotes the formation of planar membrane sheets. Structural and mutagenesis analyses reveal that the BAR domain of Pinkbar has a relatively flat lipid-binding interface and that it assembles into sheet-like oligomers in crystals and in solution, which may explain its unique membrane-deforming activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
McMahon, H.T. & Gallop, J.L. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596 (2005).
Campelo, F., Fabrikant, G., McMahon, H.T. & Kozlov, M.M. Modeling membrane shaping by proteins: focus on EHD2 and N-BAR domains. FEBS Lett. 584, 1830–1839 (2010).
Frost, A., Unger, V.M. & De Camilli, P. The BAR domain superfamily: membrane-molding macromolecules. Cell 137, 191–196 (2009).
Suetsugu, S., Toyooka, K. & Senju, Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin. Cell Dev. Biol. 21, 340–349 (2010).
Takei, K., Slepnev, V.I., Haucke, V. & De Camilli, P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat. Cell Biol. 1, 33–39 (1999).
Peter, B.J. et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303, 495–499 (2004).
Aspenström, P.A. Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr. Biol. 7, 479–487 (1997).
Itoh, T. et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev. Cell 9, 791–804 (2005).
Tsujita, K. et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J. Cell Biol. 172, 269–279 (2006).
Shimada, A. et al. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129, 761–772 (2007).
Henne, W.M. et al. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure 15, 839–852 (2007).
Guerrier, S. et al. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004 (2009).
Frost, A. et al. Structural basis of membrane invagination by F-BAR domains. Cell 132, 807–817 (2008).
Millard, T.H. et al. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO J. 24, 240–250 (2005).
Lee, S.H. et al. Structural basis for the actin-binding function of missing-in-metastasis. Structure 15, 145–155 (2007).
Mattila, P.K., Salminen, M., Yamashiro, T. & Lappalainen, P. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J. Biol. Chem. 278, 8452–8459 (2003).
Woodings, J.A., Sharp, S.J. & Machesky, L.M. MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta. Biochem. J. 371, 463–471 (2003).
Saarikangas, J. et al. ABBA regulates plasma-membrane and actin dynamics to promote radial glia extension. J. Cell Sci. 121, 1444–1454 (2008).
Scita, G., Confalonieri, S., Lappalainen, P. & Suetsugu, S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 18, 52–60 (2008).
Suetsugu, S. et al. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J. Biol. Chem. 281, 35347–35358 (2006).
Mattila, P.K. et al. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J. Cell Biol. 176, 953–964 (2007).
Saarikangas, J. et al. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr. Biol. 19, 95–107 (2009).
Bhatia, V.K. et al. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 28, 3303–3314 (2009).
Suetsugu, S. et al. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP3, and Rac. J. Cell Biol. 173, 571–585 (2006).
Disanza, A. et al. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat. Cell Biol. 8, 1337–1347 (2006).
Lim, K.B. et al. The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 283, 20454–20472 (2008).
Millard, T.H., Dawson, J. & Machesky, L.M. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J. Cell Sci. 120, 1663–1672 (2007).
Kim, M.H. et al. Enhanced NMDA receptor-mediated synaptic transmission, enhanced long-term potentiation, and impaired learning and memory in mice lacking IRSp53. J. Neurosci. 29, 1586–1595 (2009).
Sawallisch, C. et al. The insulin receptor substrate of 53 kDa (IRSp53) limits hippocampal synaptic plasticity. J. Biol. Chem. 284, 9225–9236 (2009).
Chauhan, B.K. et al. Cdc42- and IRSp53-dependent contractile filopodia tether presumptive lens and retina to coordinate epithelial invagination. Development 136, 3657–3667 (2009).
Quinones, G.A., Jin, J. & Oro, A.E. I-BAR protein antagonism of endocytosis mediates directional sensing during guided cell migration. J. Cell Biol. 189, 353–367 (2010).
Wu, C. et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130 (2009).
Hidalgo, I.J., Raub, T.J. & Borchardt, R.T. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 96, 736–749 (1989).
Gallop, J.L. et al. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 25, 2898–2910 (2006).
Honda, Y. et al. Thermal unfolding of chitosanase from Streptomyces sp. N174: role of tryptophan residues in the protein structure stabilization. Biochim. Biophys. Acta 1429, 365–376 (1999).
Clark, E.H., East, J.M. & Lee, A.G. The role of tryptophan residues in an integral membrane protein: diacylglycerol kinase. Biochemistry 42, 11065–11073 (2003).
Morimoto, S. et al. Rab13 mediates the continuous endocytic recycling of occludin to the cell surface. J. Biol. Chem. 280, 2220–2228 (2005).
Yamamura, R., Nishimura, N., Nakatsuji, H., Arase, S. & Sasaki, T. The interaction of JRAB/MICAL-L2 with Rab8 and Rab13 coordinates the assembly of tight junctions and adherens junctions. Mol. Biol. Cell 19, 971–983 (2008).
Massari, S. et al. LIN7 mediates the recruitment of IRSp53 to tight junctions. Traffic 10, 246–257 (2009).
Saarikangas, J. et al. Missing-in-metastasis (MIM/MTSS1) promotes actin assembly at intercellular junctions and is required for kidney epithelia integrity. J. Cell Sci. 124, 1245–1255 (2011).
Weissenhorn, W. Crystal structure of the endophilin-A1 BAR domain. J. Mol. Biol. 351, 653–661 (2005).
Reider, A. et al. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 28, 3103–3116 (2009).
Stimpson, H.E., Toret, C.P., Cheng, A.T., Pauly, B.S. & Drubin, D.G. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol. Biol. Cell 20, 4640–4651 (2009).
Henne, W.M. et al. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science 328, 1281–1284 (2010).
Peränen, J., Rikkonen, M., Hyvonen, M. & Kaariainen, L. T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373 (1996).
Zwart, P.H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Mastronarde, D.N. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343–352 (1997).
Kremer, J.R., Mastronarde, D.N. & McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Uchiyama, K. et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J. Cell Biol. 159, 855–866 (2002).
Acknowledgements
This work was supported by the US National Institutes of Health (NIH), National Institute of Mental Health grant MH087950 to R.D. and grants from the Finnish Cancer Foundation and Academy of Finland to P.L. H.Z. was supported by the Academy of Finland, and A.P. and J.S. were supported by fellowships from Viikki Graduate School in Biosciences (VGSB) and Helsinki Graduate Program in Biotechnology and Molecular Biology (GPBM), respectively. Use of the IMCA-CAT beamline 17-BM was supported by the Industrial Macromolecular Crystallography Association through a contract with the Hauptman-Woodward Medical Research Institute. The Advanced Photon Source was supported by Department of Energy Contract W-31-109-Eng-38. We acknowledge A.-L. Nyfors, A. Salminen and A. Strandell for excellent technical assistance.
Author information
Authors and Affiliations
Contributions
A.P., M.B., H.Z., J.S., G.R., M.J., J.H., E.V.K., H.V. and R.D. performed the experiments. A.P., J.P., E.J., M.S., E.I., R.D. and P.L designed and supervised the project. A.P., R.D. and P.L. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Methods (PDF 2383 kb)
Rights and permissions
About this article
Cite this article
Pykäläinen, A., Boczkowska, M., Zhao, H. et al. Pinkbar is an epithelial-specific BAR domain protein that generates planar membrane structures. Nat Struct Mol Biol 18, 902–907 (2011). https://doi.org/10.1038/nsmb.2079
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2079
This article is cited by
-
BAIAP2L2 is a novel prognostic biomarker related to migration and invasion of HCC and associated with cuprotosis
Scientific Reports (2023)
-
Whole exome analysis of patients in Japan with hearing loss reveals high heterogeneity among responsible and novel candidate genes
Orphanet Journal of Rare Diseases (2022)
-
BAIAP2L2 facilitates the malignancy of prostate cancer (PCa) via VEGF and apoptosis signaling pathways
Genes & Genomics (2021)
-
Superresolution microscopy reveals distinct localisation of full length IRSp53 and its I-BAR domain protein within filopodia
Scientific Reports (2019)
-
Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7
Nature Communications (2019)