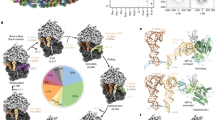
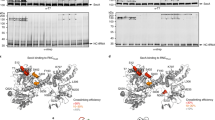
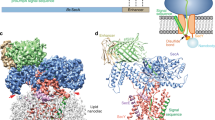
Abstract
The ubiquitous SecY–Sec61 complex translocates nascent secretory proteins across cellular membranes and integrates membrane proteins into lipid bilayers. Several structures of mostly detergent-solubilized Sec complexes have been reported. Here we present a single-particle cryo-EM structure of the SecYEG complex in a membrane environment, bound to a translating ribosome, at subnanometer resolution. Using the SecYEG complex reconstituted in a so-called Nanodisc, we could trace the nascent polypeptide chain from the peptidyltransferase center into the membrane. The reconstruction allowed for the identification of ribosome–lipid interactions. The rRNA helix 59 (H59) directly contacts the lipid surface and appears to modulate the membrane in immediate vicinity to the proposed lateral gate of the protein-conducting channel (PCC). On the basis of our map and molecular dynamics simulations, we present a model of a signal anchor–gated PCC in the membrane.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rapoport, T.A. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 (2007).
Halic, M. & Beckmann, R. The signal recognition particle and its interactions during protein targeting. Curr. Opin. Struct. Biol. 15, 116–125 (2005).
Van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004).
Zimmer, J., Nam, Y. & Rapoport, T.A. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 (2008).
Tsukazaki, T. et al. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455, 988–991 (2008).
Egea, P.F. & Stroud, R.M. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. USA 107, 17182–17187 (2010).
du Plessis, D.J., Berrelkamp, G., Nouwen, N. & Driessen, A.J. The lateral gate of SecYEG opens during protein translocation. J. Biol. Chem. 284, 15805–15814 (2009).
Osborne, A.R. & Rapoport, T.A. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell 129, 97–110 (2007).
Mitra, K. et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 (2005).
Menetret, J.-F. et al. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol. Cell 28, 1083–1092 (2007).
Menetret, J.-F. et al. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure 16, 1126–1137 (2008).
Kalies, K.U., Stokes, V. & Hartmann, E. A single Sec61 complex functions as a protein-conducting channel. Biochim. Biophys. Acta 1783, 2375–2383 (2008).
Becker, T. et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 (2009).
Gumbart, J., Trabuco, L.G., Schreiner, E., Villa, E. & Schulten, K. Regulation of the protein–conducting channel by a bound ribosome. Structure 17, 1453–1464 (2009).
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001).
Beckmann, R. et al. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science 278, 2123–2126 (1997).
Menetret, J.-F. et al. Architecture of the ribosome-channel complex derived from native membranes. J. Mol. Biol. 348, 445–457 (2005).
Morgan, D.G., Menetret, J.F., Neuhof, A., Rapoport, T.A. & Akey, C.W. Structure of the mammalian ribosome-channel complex at 17A resolution. J. Mol. Biol. 324, 871–886 (2002).
Menetret, J.-F. et al. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell 6, 1219–1232 (2000).
Matlack, K.E., Plath, K., Misselwitz, B. & Rapoport, T.A. Protein transport by purified yeast Sec complex and Kar2p without membranes. Science 277, 938–941 (1997).
Raunser, S. & Walz, T. Electron crystallography as a technique to study the structure on membrane proteins in a lipidic environment. Annu. Rev. Biophys. 38, 89–105 (2009).
Wang, L. & Sigworth, F.J. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature 461, 292–295 (2009).
Alami, M., Dalal, K., Lelj-Garolla, B., Sligar, S.G. & Duong, F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26, 1995–2004 (2007).
Shih, A.Y. et al. Molecular modeling of the structural properties and formation of high-density lipoprotein particles. in Current Topics in Membranes: Computational Modeling of Membrane Bilayers (ed. Feller, S.) 313–342 (Elsevier, 2008).
Halic, M. et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444, 507–511 (2006).
Urbanus, M.L. et al. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529 (2001).
Hamman, B.D., Chen, J.C., Johnson, E.E. & Johnson, A.E. The aqueous pore through the translocon has a diameter of 40–60 Å during cotranslational protein translocation at the ER membrane. Cell 89, 535–544 (1997).
Trabuco, L.G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008).
Cockburn, J.J. et al. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432, 122–125 (2004).
Laurinmaki, P.A., Huiskonen, J.T., Bamford, D.H. & Butcher, S.J. Membrane proteins modulate the bilayer curvature in the bacterial virus Bam35. Structure 13, 1819–1828 (2005).
Tilley, S.J., Orlova, E.V., Gilbert, R.J., Andrew, P.W. & Saibil, H.R. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell 121, 247–256 (2005).
Wu, Z. et al. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat. Struct. Mol. Biol. 14, 861–868 (2007).
Borhani, D.W., Rogers, D.P., Engler, J.A. & Brouillette, C.G. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc. Natl. Acad. Sci. USA 94, 12291–12296 (1997).
Breyton, C., Haase, W., Rapoport, T.A., Kuhlbrandt, W. & Collinson, I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665 (2002).
Collinson, I. The structure of the bacterial protein translocation complex SecYEG. Biochem. Soc. Trans. 33, 1225–1230 (2005).
Chiba, K., Mori, H. & Ito, K. Roles of the C-terminal end of SecY in protein translocation and viability of Escherichia coli. J. Bacteriol. 184, 2243–2250 (2002).
Murphy, C.K. & Beckwith, J. Residues essential for the function of SecE, a membrane component of the Escherichia coli secretion apparatus, are located in a conserved cytoplasmic region. Proc. Natl. Acad. Sci. USA 91, 2557–2561 (1994).
Houben, E.N., Zarivach, R., Oudega, B. & Luirink, J. Early encounters of a nascent membrane protein: specificity and timing of contacts inside and outside the ribosome. J. Cell Biol. 170, 27–35 (2005).
Bornemann, T., Jockel, J., Rodnina, M.V. & Wintermeyer, W. Signal sequence–independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15, 494–499 (2008).
Gumbart, J. & Schulten, K. Structural determinants of lateral gate opening in the protein translocon. Biochemistry 46, 11147–11157 (2007).
Scotti, P.A. et al. The Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 (2000).
van der Laan, M., Houben, E.N., Nouwen, N., Luirink, J. & Driessen, A.J. Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Rep. 2, 519–523 (2001).
Michanek, A., Kristen, N., Hook, F., Nylander, T. & Sparr, E. RNA and DNA interactions with zwitterionic and charged lipid membranes—a DSC and QCM-D study. Biochim. Biophys. Acta 4, 829–838 (1798).
Khvorova, A., Kwak, Y.G., Tamkun, M., Majerfeld, I. & Yarus, M. RNAs that bind and change the permeability of phospholipid membranes. Proc. Natl. Acad. Sci. USA 96, 10649–10654 (1999).
Janas, T. & Yarus, M. Specific RNA binding to ordered phospholipid bilayers. Nucleic Acids Res. 34, 2128–2136 (2006).
Marty, R., N'Soukpoe-Kossi, C.N., Charbonneau, D.M., Kreplak, L. & Tajmir-Riahi, H.A. Structural characterization of cationic lipid-tRNA complexes. Nucleic Acids Res. 37, 5197–5207 (2009).
Jaud, S. et al. Insertion of short transmembrane helices by the Sec61 translocon. Proc. Natl. Acad. Sci. USA 106, 11588–11593 (2009).
von Heijne, G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 5, 3021–3027 (1986).
van der Does, C., de Keyzer, J., van der Laan, M. & Driessen, A.J. Reconstitution of purified bacterial preprotein translocase in liposomes. Methods Enzymol. 372, 86–98 (2003).
Denisov, I.G., Grinkova, Y.V., Lazarides, A.A. & Sligar, S.G. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126, 3477–3487 (2004).
Wagenknecht, T., Grassucci, R. & Frank, J. Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J. Mol. Biol. 199, 137–147 (1988).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Chen, J.Z. & Grigorieff, N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 157, 168–173 (2007).
Berk, V., Zhang, W., Pai, R.D. & Cate, J.H. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. USA 103, 15830–15834 (2006).
Seidelt, B. et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326, 1412–1415 (2009).
Söding, J., Biegert, A. & Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–8 (2005).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Bostina, M., Mohsin, B., Kühlbrandt, W. & Collinson, I. Atomic model of the E. coli membrane-bound protein translocation complex SecYEG. J. Mol. Biol. 352, 1035–1043 (2005).
Breyton, C., Haase, W., Rapoport, T.A., Kühlbrandt, W. & Collinson, I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665 (2002).
Acknowledgements
We thank B. Seidelt for help with the RNC preparation, J.P. Armache for help with data processing, S. Feuerstein (Forschungszentrum Jülich) for providing the plasmid encoding the apolipoprotein construct, J. Buerger and C. Ungewickell for help with the electron microscopy and D. Wilson for critical discussions. This research was supported by a Boehringer Ingelheim Fonds fellowship (to J.F.), by an Alexander von Humboldt Foundation fellowship and a Human Frontiers Science Program long term fellowship (to E.O.v.d.S.), by a Dr. Klaus Römer Stiftung fellowship (to T.B.), by grants from the Deutsche Forschungsgemeinschaft SFB594 and SFB646 (to R.B.) and SFB 740 (to T.M.), by US National Institutes of Health grants P41-RR005969 and R01-GM067887 (to K.S.), by US National Science Foundation (NSF) grant PHY0822613 (to K.S.) by an Alexander von Humboldt Foundation award (to K.S.) and by the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum). Computer time for MD simulations and MDFF was provided through NSF Large Resources Allocation Committee grant MCA93S028.
Author information
Authors and Affiliations
Contributions
J.F. prepared the sample, collected the EM data, performed the 3D reconstruction and built the molecular model; J.G. did the MDFF and the MD simulations. E.O.v.d.S., S.F. and B.B. contributed to the purification of SecYEG; M.G. contributed to the data processing. T.M. and O.B. contributed to the EM data collection. T.B. contributed to model building and interpretation. All authors contributed to the study design and to writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Figures 1–12 and Supplementary Tables 1–7 (PDF 3367 kb)
Supplementary Movie
Molecular dynamics simulation (MOV 29075 kb)
Rights and permissions
About this article
Cite this article
Frauenfeld, J., Gumbart, J., Sluis, E. et al. Cryo-EM structure of the ribosome–SecYE complex in the membrane environment. Nat Struct Mol Biol 18, 614–621 (2011). https://doi.org/10.1038/nsmb.2026
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2026
This article is cited by
-
Modulating co-translational protein folding by rational design and ribosome engineering
Nature Communications (2022)
-
Molecular communication of the membrane insertase YidC with translocase SecYEG affects client proteins
Scientific Reports (2021)
-
Characterization of the Features of Water Inside the SecY Translocon
The Journal of Membrane Biology (2021)
-
Cardiolipin is required in vivo for the stability of bacterial translocon and optimal membrane protein translocation and insertion
Scientific Reports (2020)
-
Protein Transport Across the Bacterial Plasma Membrane by the Sec Pathway
The Protein Journal (2019)