Abstract
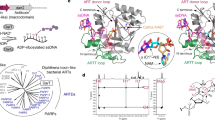
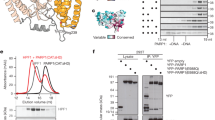
Aprataxin is a DNA deadenylase that resolves DNA 5′-AMP termini and reverses abortive DNA ligation. The crystal structures of Schizosaccharomyces pombe aprataxin Hnt3 in its apo form and in complex to dsDNA and dsDNA–AMP reveal how Hnt3 recognizes and processes 5′-adenylated DNA in a structure-specific manner. The bound DNA adopts a 5′-flap conformation that facilitates 5′-AMP access to the active site, where AMP cleavage occurs by a canonical catalytic mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Aicardi, J. et al. Ann. Neurol. 24, 497–502 (1988).
Date, H. et al. Nat. Genet. 29, 184–188 (2001).
Moreira, M.C. et al. Nat. Genet. 29, 189–193 (2001).
Ahel, I. et al. Nature 443, 713–716 (2006).
Kijas, A.W., Harris, J.L., Harris, J.M. & Lavin, M.F. J. Biol. Chem. 281, 13939–13948 (2006).
Rass, U., Ahel, I. & West, S.C. J. Biol. Chem. 282, 9469–9474 (2007).
Rass, U., Ahel, I. & West, S.C. Cell 130, 991–1004 (2007).
Caldecott, K.W. Nat. Rev. Genet. 9, 619–631 (2008).
Rass, U., Ahel, I. & West, S.C. J. Biol. Chem. 283, 33994–34001 (2008).
Sykora, P., Croteau, D.L., Bohr, V.A. & Wilson, D.M. III. Proc. Natl. Acad. Sci. USA 108, 7437–7442 (2011).
Brenner, C. Biochemistry 41, 9003–9014 (2002).
Deshpande, G.P. et al. DNA Repair (Amst.) 8, 672–679 (2009).
Lima, C.D., Klein, M.G. & Hendrickson, W.A. Science 278, 286–290 (1997).
Wolfe, S.A., Nekludova, L. & Pabo, C.O. Annu. Rev. Biophys. Biomol. Struct. 29, 183–212 (2000).
Clements, P.M. et al. DNA Repair (Amst.) 3, 1493–1502 (2004).
Becherel, O.J. et al. Nucleic Acids Res. 38, 1489–1503 (2010).
Acknowledgements
We are grateful to Y. Chen for her assistance in Biacore analysis. We thank the staff at the Shanghai Synchrotron Radiation Facility beamline 17U for assistance with data collection, and we thank our colleagues for critical comments on the manuscript. This work was funded by Chinese Ministry of Science and Technology '973' grants 2011CB910302 and 2011CB910304 to T.J. and D.-C.W.; National Natural Science Foundation of China grants 31021062 and 31025009 to T.J. and 31000330 to D.Z.; and Chinese Academy of Sciences grant KSCX2-EW-J-3 to J.D. and D.-C.W.
Author information
Authors and Affiliations
Contributions
Y.G., D.Z., J.D., T.J. and D.-C.W. designed the research; Y.G., D.Z., J.D., X.R. and C.-N.D. carried out the experiments; Y.G., D.Z., J.D., T.J. and D.-C.W. analyzed the data and wrote the paper; and all authors contributed to editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1, Supplementary Discussion and Supplementary Methods (PDF 2875 kb)
Rights and permissions
About this article
Cite this article
Gong, Y., Zhu, D., Ding, J. et al. Crystal structures of aprataxin ortholog Hnt3 reveal the mechanism for reversal of 5′-adenylated DNA. Nat Struct Mol Biol 18, 1297–1299 (2011). https://doi.org/10.1038/nsmb.2145
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2145
This article is cited by
-
Structural biology: How to mend a broken DNA
Nature China (2012)