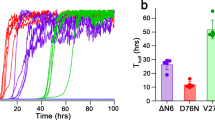
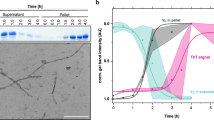
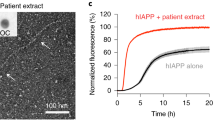
Abstract
β2-microglobulin (β2m) is the light chain of the type I major histocompatibility complex. It deposits as amyloid fibrils within joints during long-term hemodialysis treatment. Despite the devastating effects of dialysis-related amyloidosis, full understanding of how fibrils form from soluble β2m remains elusive. Here we show that β2m can oligomerize and fibrillize via three-dimensional domain swapping. Isolating a covalently bound, domain-swapped dimer from β2m oligomers on the pathway to fibrils, we were able to determine its crystal structure. The hinge loop that connects the swapped domain to the core domain includes the fibrillizing segment LSFSKD, whose atomic structure we also determined. The LSFSKD structure reveals a class 5 steric zipper, akin to other amyloid spines. The structures of the dimer and the zipper spine fit well into an atomic model for this fibrillar form of β2m, which assembles slowly under physiological conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Westermark, P. et al. Amyloid: toward terminology clarification. Report from the Nomenclature Committee of the International Society of Amyloidosis. Amyloid 12, 1–4 (2005).
Chiti, F. & Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Makin, O.S. & Serpell, L.C. Structures for amyloid fibrils. FEBS J. 272, 5950–5961 (2005).
Nelson, R. et al. Structure of the cross-β spine of amyloid-like fibrils. Nature 435, 773–778 (2005).
Sawaya, M.R. et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447, 453–457 (2007).
Tycko, R. Molecular structure of amyloid fibrils: insights from solid-state NMR. Q. Rev. Biophys. 39, 1–55 (2006).
Margittai, M. & Langen, R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q. Rev. Biophys. 41, 265–297 (2008).
Glabe, C.G. Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol. Aging 27, 570–575 (2006).
Bucciantini, M. et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416, 507–511 (2002).
Wahlbom, M. et al. Fibrillogenic oligomers of human cystatin C are formed by propagated domain swapping. J. Biol. Chem. 282, 18318–18326 (2007).
Guo, Z. & Eisenberg, D. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc. Natl. Acad. Sci. USA 103, 8042–8047 (2006).
Knaus, K.J. et al. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 8, 770–774 (2001).
Janowski, R. et al. Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Biol. 8, 316–320 (2001).
Gronenborn, A.M. Protein acrobatics in pairs—dimerization via domain swapping. Curr. Opin. Struct. Biol. 19, 39–49 (2009).
Bennett, M.J., Sawaya, M.R. & Eisenberg, D. Deposition diseases and 3D domain swapping. Structure 14, 811–824 (2006).
Becker, J.W. & Reeke, G.N. Jr. Three-dimensional structure of β2-microglobulin. Proc. Natl. Acad. Sci. USA 82, 4225–4229 (1985).
Campistol, J.M. et al. Polymerization of normal and intact β2-microglobulin as the amyloidogenic protein in dialysis-amyloidosis. Kidney Int. 50, 1262–1267 (1996).
McParland, V.J., Kalverda, A.P., Homans, S.W. & Radford, S.E. Structural properties of an amyloid precursor of β2-microglobulin. Nat. Struct. Biol. 9, 326–331 (2002).
McParland, V.J. et al. Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry 39, 8735–8746 (2000).
Eakin, C.M., Berman, A.J. & Miranker, A.D. A native to amyloidogenic transition regulated by a backbone trigger. Nat. Struct. Mol. Biol. 13, 202–208 (2006).
Yamaguchi, K., Naiki, H. & Goto, Y. Mechanism by which the amyloid-like fibrils of a β2-microglobulin fragment are induced by fluorine-substituted alcohols. J. Mol. Biol. 363, 279–288 (2006).
Myers, S.L. et al. A systematic study of the effect of physiological factors on β2-microglobulin amyloid formation at neutral pH. Biochemistry 45, 2311–2321 (2006).
Heegaard, N.H. β2-microglobulin: from physiology to amyloidosis. Amyloid 16, 151–173 (2009).
Jahn, T.R., Parker, M.J., Homans, S.W. & Radford, S.E. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 13, 195–201 (2006).
Esposito, G. et al. Removal of the N-terminal hexapeptide from human β2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 9, 831–845 (2000).
Platt, G.W., Routledge, K.E., Homans, S.W. & Radford, S.E. Fibril growth kinetics reveal a region of β2-microglobulin important for nucleation and elongation of aggregation. J. Mol. Biol. 378, 251–263 (2008).
Iwata, K. et al. 3D structure of amyloid protofilaments of β2-microglobulin fragment probed by solid-state NMR. Proc. Natl. Acad. Sci. USA 103, 18119–18124 (2006).
Ivanova, M.I., Sawaya, M.R., Gingery, M., Attinger, A. & Eisenberg, D. An amyloid-forming segment of β2-microglobulin suggests a molecular model for the fibril. Proc. Natl. Acad. Sci. USA 101, 10584–10589 (2004).
Blaho, D.V. & Miranker, A.D. Delineating the conformational elements responsible for Cu2+-induced oligomerization of β2-microglobulin. Biochemistry 48, 6610–6617 (2009).
Calabrese, M.F., Eakin, C.M., Wang, J.M. & Miranker, A.D. A regulatable switch mediates self-association in an immunoglobulin fold. Nat. Struct. Mol. Biol. 15, 965–971 (2008).
Katou, H. et al. The role of disulfide bond in the amyloidogenic state of β2-microglobulin studied by heteronuclear NMR. Protein Sci. 11, 2218–2229 (2002).
Smith, D.P. & Radford, S.E. Role of the single disulphide bond of β2-microglobulin in amyloidosis in vitro. Protein Sci. 10, 1775–1784 (2001).
Eakin, C.M., Attenello, F.J., Morgan, C.J. & Miranker, A.D. Oligomeric assembly of native-like precursors precedes amyloid formation by β2-microglobulin. Biochemistry 43, 7808–7815 (2004).
Trinh, C.H., Smith, D.P., Kalverda, A.P., Phillips, S.E. & Radford, S.E. Crystal structure of monomeric human β2-microglobulin reveals clues to its amyloidogenic properties. Proc. Natl. Acad. Sci. USA 99, 9771–9776 (2002).
Khan, A.R., Baker, B.M., Ghosh, P., Biddison, W.E. & Wiley, D.C. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. Immunol. 164, 6398–6405 (2000).
Rousseau, F., Schymkowitz, J.W., Wilkinson, H.R. & Itzhaki, L.S. Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc. Natl. Acad. Sci. USA 98, 5596–5601 (2001).
Sambashivan, S., Liu, Y., Sawaya, M.R., Gingery, M. & Eisenberg, D. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature 437, 266–269 (2005).
Lee, S. & Eisenberg, D. Seeded conversion of recombinant prion protein to a disulfide-bonded oligomer by a reduction-oxidation process. Nat. Struct. Biol. 10, 725–730 (2003).
Biancalana, M., Makabe, K. & Koide, S. Minimalist design of water-soluble cross-β architecture. Proc. Natl. Acad. Sci. USA 107, 3469–3474 (2010).
Jahn, T.R. et al. The common architecture of cross-β amyloid. J. Mol. Biol. 395, 717–727 (2010).
Hogg, P.J. Disulfide bonds as switches for protein function. Trends Biochem. Sci. 28, 210–214 (2003).
Nilsson, M. et al. Prevention of domain swapping inhibits dimerization and amyloid fibril formation of cystatin C: use of engineered disulfide bridges, antibodies, and carboxymethylpapain to stabilize the monomeric form of cystatin C. J. Biol. Chem. 279, 24236–24245 (2004).
Fändrich, M., Meinhardt, J. & Grigorieff, N. Structural polymorphism of Alzheimer Aβ and other amyloid fibrils. Prion 3, 89–93 (2009).
Kodali, R. & Wetzel, R. Polymorphism in the intermediates and products of amyloid assembly. Curr. Opin. Struct. Biol. 17, 48–57 (2007).
Goldsbury, C.S. et al. Polymorphic fibrillar assembly of human amylin. J. Struct. Biol. 119, 17–27 (1997).
Paravastu, A.K., Leapman, R.D., Yau, W.M. & Tycko, R. Molecular structural basis for polymorphism in Alzheimer′s β-amyloid fibrils. Proc. Natl. Acad. Sci. USA 105, 18349–18354 (2008).
Wiltzius, J.J. et al. Molecular mechanisms for protein-encoded inheritance. Nat. Struct. Mol. Biol. 16, 973–978 (2009).
Ladner, C.L. et al. Stacked sets of parallel, in-register β-strands of β2-microglobulin in amyloid fibrils revealed by site-directed spin labeling and chemical labeling. J. Biol. Chem. 285, 17137–17147 (2010).
Yamamoto, K. et al. Thiol compounds inhibit the formation of amyloid fibrils by β2-microglobulin at neutral pH. J. Mol. Biol. 376, 258–268 (2008).
Chen, Y. & Dokholyan, N.V. A single disulfide bond differentiates aggregation pathways of β2-microglobulin. J. Mol. Biol. 354, 473–482 (2005).
Stoppini, M. et al. Proteomics of β2-microglobulin amyloid fibrils. Biochim. Biophys. Acta 1753, 23–33 (2005).
Bellotti, V. β2-microglobulin can be refolded into a native state from ex vivo amyloid fibrils. Eur. J. Biochem. 258, 61–67 (1998).
Platt, G.W. & Radford, S.E. Glimpses of the molecular mechanisms of β2-microglobulin fibril formation in vitro: aggregation on a complex energy landscape. FEBS Lett. 583, 2623–2629 (2009).
Gorevic, P.D. et al. Beta-2 microglobulin is an amyloidogenic protein in man. J. Clin. Invest. 76, 2425–2429 (1985).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Acknowledgements
We thank the Northeastern Collaborative Access Team facility at the Advanced Photon Source at Argonne National Laboratory for beam time and collection assistance, and S. Radford, M. Bennett and Z. Guo for discussion. This work was supported by the US National Institutes of Health, the US Department of Energy Biological and Environmental Research program and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
C.L. designed the research, with advice from D.E., and carried out all the experiments; M.R.S. calculated and built the fibril models; C.L. wrote the paper; all authors discussed the results and revised the manuscript; D.E. supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Methods (PDF 926 kb)
Rights and permissions
About this article
Cite this article
Liu, C., Sawaya, M. & Eisenberg, D. β2-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat Struct Mol Biol 18, 49–55 (2011). https://doi.org/10.1038/nsmb.1948
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1948
This article is cited by
-
Assembly of platforms for signal transduction in the new era: dimerization, helical filament assembly, and beyond
Experimental & Molecular Medicine (2020)
-
Probing Medin Monomer Structure and its Amyloid Nucleation Using 13C-Direct Detection NMR in Combination with Structural Bioinformatics
Scientific Reports (2017)
-
A covalent homodimer probing early oligomers along amyloid aggregation
Scientific Reports (2015)
-
A review on protein oligomerization process
International Journal of Precision Engineering and Manufacturing (2015)