Abstract
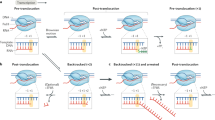
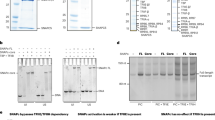
The transition from transcription initiation to elongation at the HIV-1 promoter is controlled by Tat, which recruits P-TEFb to TAR RNA to phosphorylate RNA polymerase II. It has long been unclear why the HIV-1 promoter is incompetent for elongation. We report that P-TEFb is recruited to the promoter in a catalytically inactive state bound to the inhibitory 7SK small nuclear ribonucleoprotein (snRNP), thereby preventing elongation. It also has long been believed that TAR functions to recruit Tat to the promoter, but we find that Tat is recruited to the DNA template before TAR is synthesized. We propose that TAR binds Tat and P-TEFb as it emerges on the nascent transcript, competitively displacing the inhibitory 7SK snRNP and activating the P-TEFb kinase. Recruitment of an inhibitory snRNP complex at an early stage in the transcription cycle provides a new paradigm for controlling gene expression with a noncoding RNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sims, R.J. III, Belotserkovskaya, R. & Reinberg, D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18, 2437–2468 (2004).
Feinberg, M.B., Baltimore, D. & Frankel, A.D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 88, 4045–4049 (1991).
Laspia, M.F., Rice, A.P. & Mathews, M.B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59, 283–292 (1989).
Garber, M.E. et al. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12, 3512–3527 (1998).
Raha, T., Cheng, S.W. & Green, M.R. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 3, e44 (2005).
Zhang, Z., Klatt, A., Gilmour, D.S. & Henderson, A.J. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem. 282, 16981–16988 (2007).
Core, L.J. & Lis, J.T. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319, 1791–1792 (2008).
Peterlin, B.M. & Price, D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305 (2006).
Wei, P., Garber, M.E., Fang, S.M., Fischer, W.H. & Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462 (1998).
Zhou, Q. & Yik, J.H. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70, 646–659 (2006).
Komarnitsky, P., Cho, E.J. & Buratowski, S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14, 2452–2460 (2000).
Kim, Y.K., Bourgeois, C.F., Isel, C., Churcher, M.J. & Karn, J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol. Cell. Biol. 22, 4622–4637 (2002).
Ahn, S.H., Kim, M. & Buratowski, S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13, 67–76 (2004).
Phatnani, H.P. & Greenleaf, A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 (2006).
Gomes, N.P. et al. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20, 601–612 (2006).
Buratowski, S. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36, 541–546 (2009).
Jang, M.K. et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534 (2005).
Yang, Z. et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545 (2005).
Yik, J.H. et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12, 971–982 (2003).
Jeronimo, C. et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27, 262–274 (2007).
D'Orso, I., Grunwell, J.R., Nakamura, R.L., Das, C. & Frankel, A.D. Targeting tat inhibitors in the assembly of human immunodeficiency virus type 1 transcription complexes. J. Virol. 82, 9492–9504 (2008).
Pan, G., Aso, T. & Greenblatt, J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J. Biol. Chem. 272, 24563–24571 (1997).
Barboric, M. et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35, 2003–2012 (2007).
Sedore, S.C. et al. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 35, 4347–4358 (2007).
Bres, V., Gomes, N., Pickle, L. & Jones, K.A. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 19, 1211–1226 (2005).
Bres, V., Yoshida, T., Pickle, L. & Jones, K.A. SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol. Cell 36, 75–87 (2009).
Keen, N.J., Gait, M.J. & Karn, J. Human immunodeficiency virus type-1 Tat is an integral component of the activated transcription-elongation complex. Proc. Natl. Acad. Sci. USA 93, 2505–2510 (1996).
Berkhout, B., Gatignol, A., Rabson, A.B. & Jeang, K.T. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell 62, 757–767 (1990).
Kamine, J., Subramanian, T. & Chinnadurai, G. Sp1-dependent activation of a synthetic promoter by human immunodeficiency virus type 1 Tat protein. Proc. Natl. Acad. Sci. USA 88, 8510–8514 (1991).
Krueger, B.J. et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 36, 2219–2229 (2008).
Isel, C. & Karn, J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J. Mol. Biol. 290, 929–941 (1999).
Ping, Y.H. & Rana, T.M. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J. Biol. Chem. 274, 7399–7404 (1999).
Zhou, M. et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20, 5077–5086 (2000).
Zhou, C. & Rana, T.M. A bimolecular mechanism of HIV-1 Tat protein interaction with RNA polymerase II transcription elongation complexes. J. Mol. Biol. 320, 925–942 (2002).
Garcia-Martinez, L.F., Ivanov, D. & Gaynor, R.B. Association of Tat with purified HIV-1 and HIV-2 transcription preinitiation complexes. J. Biol. Chem. 272, 6951–6958 (1997).
Glover-Cutter, K., Kim, S., Espinosa, J. & Bentley, D.L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 15, 71–78 (2008).
Niranjanakumari, S., Lasda, E., Brazas, R. & Garcia-Blanco, M.A. Reversible crosslinking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26, 182–190 (2002).
Czudnochowski, N., Vollmuth, F., Baumann, S., Vogel-Bachmayr, K. & Geyer, M. Specificity of Hexim1 and Hexim2 complex formation with cyclin T1/T2, importin alpha and 7SK snRNA. J. Mol. Biol. 395, 28–41 (2010).
Schulte, A. et al. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 280, 24968–24977 (2005).
Berkhout, B., Silverman, R.H. & Jeang, K.T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59, 273–282 (1989).
Kamine, J. & Chinnadurai, G. Synergistic activation of the human immunodeficiency virus type 1 promoter by the viral Tat protein and cellular transcription factor Sp1. J. Virol. 66, 3932–3936 (1992).
Yedavalli, V.S., Benkirane, M. & Jeang, K.T. Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J. Biol. Chem. 278, 6404–6410 (2003).
Montanuy, I., Torremocha, R., Hernandez-Munain, C. & Sune, C. Promoter influences transcription elongation: TATA-box element mediates the assembly of processive transcription complexes responsive to cyclin-dependent kinase 9. J. Biol. Chem. 283, 7368–7378 (2008).
Cujec, T.P. et al. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol. Cell. Biol. 17, 1817–1823 (1997).
Fraldi, A. et al. Inhibition of Tat activity by the HEXIM1 protein. Retrovirology 2, 42 (2005).
Southgate, C., Zapp, M.L. & Green, M.R. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature 345, 640–642 (1990).
Selby, M.J. & Peterlin, B.M. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell 62, 769–776 (1990).
Smith, C.A., Calabro, V. & Frankel, A.D. An RNA-binding chameleon. Mol. Cell 6, 1067–1076 (2000).
D'Orso, I. & Frankel, A.D. Tat acetylation modulates assembly of a viral-host RNA-protein transcription complex. Proc. Natl. Acad. Sci. USA 106, 3101–3106 (2009).
Casse, C., Giannoni, F., Nguyen, V.T., Dubois, M.F. & Bensaude, O. The transcriptional inhibitors, actinomycin D and α-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J. Biol. Chem. 274, 16097–16106 (1999).
Biglione, S. et al. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology 4, 47 (2007).
Ujvari, A. & Luse, D.S. Newly initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J. Biol. Chem. 279, 49773–49779 (2004).
Kornblihtt, A.R., de la Mata, M., Fededa, J.P., Munoz, M.J. & Nogues, G. Multiple links between transcription and splicing. RNA 10, 1489–1498 (2004).
Fong, Y.W. & Zhou, Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414, 929–933 (2001).
Young, T.M., Tsai, M., Tian, B., Mathews, M.B. & Pe'ery, T. Cellular mRNA activates transcription elongation by displacing 7SK RNA. PLoS One 2, e1010 (2007).
Southgate, C.D. & Green, M.R. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 5, 2496–2507 (1991).
Lis, J.T., Mason, P., Peng, J., Price, D.H. & Werner, J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14, 792–803 (2000).
Sobhian, B. et al. HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP in vivo. Mol. Cell 38, 439–451 (2010).
Van Herreweghe, E. et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 26, 3570–3580 (2007).
Barrandon, C., Bonnet, F., Nguyen, V.T., Labas, V. & Bensaude, O. The transcription-dependent dissociation of P-TEFb-HEXIM1–7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 27, 6996–7006 (2007).
Robert, F., Blanchette, M., Maes, O., Chabot, B. & Coulombe, B. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem. 277, 9302–9306 (2002).
Acknowledgements
We thank K. Yamamoto, P. Walter, C. Guthrie, R. Andino, J. Gross and N. Krogan for comments, J. Greenblatt (Univ. of Toronto) for reagents, members of the Frankel and Yamamoto laboratory for discussions and the reviewers for their insightful comments. This work was initially supported by an amfAR Mathilde Krim fellowship in Basic Biomedical Research (106988-43-RFNT) and a US National Institutes of Health K99/R00 grant A112185 to I.D. as well as US National Institutes of Health grants AI29135 and P50 GM082250 (HARC Center) to A.D.F.
Author information
Authors and Affiliations
Contributions
I.D. and A.D.F. designed research; I.D. performed research; I.D. and A.D.F. analyzed data and wrote the paper
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1–3 and Supplementary Methods (PDF 2208 kb)
Rights and permissions
About this article
Cite this article
D'Orso, I., Frankel, A. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol 17, 815–821 (2010). https://doi.org/10.1038/nsmb.1827
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1827
This article is cited by
-
Stochastic pausing at latent HIV-1 promoters generates transcriptional bursting
Nature Communications (2021)
-
CDK9 keeps RNA polymerase II on track
Cellular and Molecular Life Sciences (2021)
-
Cellular RelB interacts with the transactivator Tat and enhance HIV-1 expression
Retrovirology (2018)
-
PJA2 ubiquitinates the HIV-1 Tat protein with atypical chain linkages to activate viral transcription
Scientific Reports (2017)
-
HIV-1 transcription and latency: an update
Retrovirology (2013)