Abstract
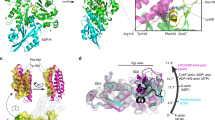
Capping protein (CP) regulates actin dynamics by binding the barbed ends of actin filaments. Removal of CP may be one means to harness actin polymerization for processes such as cell movement and endocytosis. Here we structurally and biochemically investigated a CP interaction (CPI) motif present in the otherwise unrelated proteins CARMIL and CD2AP. The CPI motif wraps around the stalk of the mushroom-shaped CP at a site distant from the actin-binding interface, which lies on the top of the mushroom cap. We propose that the CPI motif may act as an allosteric modulator, restricting CP to a low-affinity, filament-binding conformation. Structure-based sequence alignments extend the CPI motif–containing family to include CIN85, CKIP-1, CapZIP and a relatively uncharacterized protein, WASHCAP (FAM21). Peptides comprising these CPI motifs are able to inhibit CP and to uncap CP-bound actin filaments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
20 December 2011
In the version of this article initially published online, the captions for the supplementary videos were missing. The error has been corrected in the HTML version of the article.
References
Kovar, D.R., Wu, J.Q. & Pollard, T.D. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell 16, 2313–2324 (2005).
Wear, M.A., Yamashita, A., Kim, K., Maeda, Y. & Cooper, J.A. How capping protein binds the barbed end of the actin filament. Curr. Biol. 13, 1531–1537 (2003).
Cooper, J.A. & Sept, D. New insights into mechanism and regulation of actin capping protein. Int. Rev. Cell Mol. Biol. 267, 183–206 (2008).
Mejillano, M.R. et al. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 118, 363–373 (2004).
Yamashita, A., Maeda, K. & Maeda, Y. Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J. 22, 1529–1538 (2003).
Narita, A., Takeda, S., Yamashita, A. & Maeda, Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J. 25, 5626–5633 (2006).
Jung, G., Remmert, K., Wu, X., Volosky, J.M. & Hammer, J.A. III. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 153, 1479–1497 (2001).
Yang, C. et al. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev. Cell 9, 209–221 (2005).
Uruno, T., Remmert, K. & Hammer, J.A. III. CARMIL is a potent capping protein antagonist: identification of a conserved CARMIL domain that inhibits the activity of capping protein and uncaps capped actin filaments. J. Biol. Chem. 281, 10635–10650 (2006).
Fujiwara, I., Remmert, K. & Hammer, J.A. III. Direct observation of the uncapping of capping protein-capped actin filaments by CARMIL homology domain 3 (CAH3). J. Biol. Chem. 285, 2707–2720 (2010).
Canton, D.A. et al. The pleckstrin homology domain-containing protein CKIP-1 is involved in regulation of cell morphology and the actin cytoskeleton and interaction with actin capping protein. Mol. Cell. Biol. 25, 3519–3534 (2005).
Canton, D.A., Olsten, M.E., Niederstrasser, H., Cooper, J.A. & Litchfield, D.W. The role of CKIP-1 in cell morphology depends on its interaction with actin-capping protein. J. Biol. Chem. 281, 36347–36359 (2006).
Bruck, S. et al. Identification of a novel inhibitory actin-capping protein binding motif in CD2-associated protein. J. Biol. Chem. 281, 19196–19203 (2006).
Gaidos, G., Soni, S., Oswald, D.J., Toselli, P.A. & Kirsch, K.H. Structure and function analysis of the CMS/CIN85 protein family identifies actin-bundling properties and heterotypic-complex formation. J. Cell Sci. 120, 2366–2377 (2007).
Hutchings, N.J., Clarkson, N., Chalkley, R., Barclay, A.N. & Brown, M.H. Linking the T cell surface protein CD2 to the actin-capping protein CAPZ via CMS and CIN85. J. Biol. Chem. 278, 22396–22403 (2003).
Eyers, C.E. et al. The phosphorylation of CapZ-interacting protein (CapZIP) by stress-activated protein kinases triggers its dissociation from CapZ. Biochem. J. 389, 127–135 (2005).
Bhattacharya, N., Ghosh, S., Sept, D. & Cooper, J.A. Binding of myotrophin/V-1 to actin-capping protein: implications for how capping protein binds to the filament barbed end. J. Biol. Chem. 281, 31021–31030 (2006).
Kim, K. et al. Structure/function analysis of the interaction of phosphatidylinositol 4,5-bisphosphate with actin-capping protein: implications for how capping protein binds the actin filament. J. Biol. Chem. 282, 5871–5879 (2007).
Kuhn, J.R. & Pollard, T.P. Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J. Biol. Chem. 282, 28014–28024 (2007).
Cooper, J.A., Walker, S.B. & Pollard, T.P. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil. 4, 253–262 (1983).
Dikic, I. CIN85/CMS family of adaptor molecules. FEBS Lett. 529, 110–115 (2002).
Havrylov, S., Rzhepetskyy, Y., Malinowska, A., Drobot, L. & Redowicz, M.J. Proteins recruited by SH3 domains of Ruk/CIN85 adaptor identified by LC-MS/MS. Proteome Sci. 7, 21–39 (2009).
Badour, K. et al. The Wiskott-Aldrich syndrome protein acts downstream of CD2 and the CD2AP and PSTPIP1 adaptors to promote formation of the immunological synapse. Immunity 18, 141–154 (2003).
Johnson, R.I., Seppa, M.J. & Cagan, R.L. The Drosophila CD2AP/CIN85 orthologue Cindr regulates junctions and cytoskeleton dynamics during tissue patterning. J. Cell Biol. 180, 1191–1204 (2008).
Oishi, I., Kawakami, Y., Raya, A., Callol-Massot, C. & Izpisá Belmonte, J.C. Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya. Nat. Genet. 38, 1316–1322 (2006).
Hertzog, M. et al. The β-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117, 611–623 (2004).
Irobi, E. et al. Structural basis of actin sequestration by thymosin-β4: implications for WH2 proteins. EMBO J. 23, 3599–3608 (2004).
Dominguez, R. The β-thymosin/WH2 fold: multifunctionality and structure. Ann. NY Acad. Sci. 1112, 86–94 (2007).
Gomez, T.S. & Billadeau, D.D.A. FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711 (2009).
Derivery, E. et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 (2009).
Valdmanis, P.N. et al. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet. 80, 152–161 (2007).
Huang, C.Y. et al. A novel cellular protein, VPEF, facilitates Vaccinia virus penetration into HeLa cells through fluid phase endocytosis. J. Virol. 82, 7988–7999 (2008).
Hänisch, J. et al. Molecular dissection of Salmonella-induced membrane ruffling versus invasion. Cell. Microbiol. 12, 84–98 (2010).
Miyoshi, T. et al. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J. Cell Biol. 175, 947–955 (2006).
Schafer, D.A., Jennings, P.B. & Cooper, J.A. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135, 169–179 (1996).
Akin, O. & Mullins, R.D. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133, 841–851 (2008).
Soeno, Y. et al. Generation of functional β-actinin (CapZ) in an E. coli expression system. J. Muscle Res. Cell Motil. 19, 639–646 (1998).
Burtnick, L.D. et al. Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF. EMBO J. 23, 2713–2722 (2004).
Wang, H. et al. Helix straightening as an activation mechanism in the gelsolin superfamily of actin regulatory proteins. J. Biol. Chem. 284, 21265–21269 (2009).
Casella, J.F., Maack, D.J. & Lin, S. Purification and initial characterization of a protein from skeletal muscle that caps the barbed ends of actin filaments. J. Biol. Chem. 261, 10915–10921 (1986).
Rusinova, E. et al. Alexa and Oregon green dyes as fluorescence anisotropy probes for measuring protein-protein and protein-nucleic acid interactions. Anal. Biochem. 308, 18–25 (2002).
Owen, B.A., Lang, W.H. & McMurray, C.T. The nucleotide binding dynamics of human MSH2–MSH3 are lesion dependent. Nat. Struct. Mol. Biol. 16, 550–557 (2009).
Vinson, V.K., De La Cruz, E.M., Higgs, H.N. & Pollard, T.P. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry 37, 10871–10880 (1998).
Acknowledgements
We thank the Biomedical Research Council of A*STAR for support to R.C.R. and the National Synchrotron Radiation Research Center, a facility supported by the National Science Council of Taiwan for provision of beam time and assistance in data collection. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine. J.A.C. acknowledges US National Institutes of Health grant GM 38542 for support.
Author information
Authors and Affiliations
Contributions
M.H.-V., T.K., B.K. and A.T. carried out the work; A.H.A. created reagents used in the work; M.H.-V., T.K., M.L., J.A.C. and R.C.R. designed the work and interpreted the data; R.C.R. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1 and Supplementary Methods (PDF 1269 kb)
Supplementary Video 1
Uncapping of CP capped actin filaments in the presence of CBR115. Field of view, 60 × 120 µm2. Methods are found in Supplementary Methods. (AVI 1540 kb)
Supplementary Video 2
Uncapping of CP capped actin filaments in the presence of CBR37. Field of view, 60 × 120 µm2. Methods are found in Supplementary Methods. (AVI 1576 kb)
Supplementary Video 3
CP capped actin filaments in the presence of CD2AP CPI motif. Field of view, 60 × 120 µm2. Methods are found in Supplementary Methods. (AVI 1300 kb)
Supplementary Video 4
Actin filaments in the absence of CP. Field of view, 60 × 120 µm2. Methods are found in Supplementary Methods. (AVI 1175 kb)
Supplementary Video 5
CP capped actin filaments in the absence of CARMIL or CD2AP truncations. Field of view, 60 × 120 µm2. Methods are found in Supplementary Methods. (AVI 1320 kb)
Rights and permissions
About this article
Cite this article
Hernandez-Valladares, M., Kim, T., Kannan, B. et al. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat Struct Mol Biol 17, 497–503 (2010). https://doi.org/10.1038/nsmb.1792
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1792
This article is cited by
-
Twinfilin uncaps filament barbed ends to promote turnover of lamellipodial actin networks
Nature Cell Biology (2021)
-
FAM21 directs SNX27–retromer cargoes to the plasma membrane by preventing transport to the Golgi apparatus
Nature Communications (2016)
-
Formin and capping protein together embrace the actin filament in a ménage à trois
Nature Communications (2015)
-
CPI motif interaction is necessary for capping protein function in cells
Nature Communications (2015)
-
Capping protein regulators fine-tune actin assembly dynamics
Nature Reviews Molecular Cell Biology (2014)