Abstract
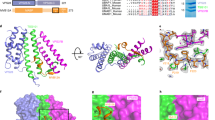
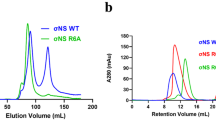
Protein folding is often mediated by molecular chaperones. Recently, a novel class of intramolecular chaperones has been identified in tailspike proteins of evolutionarily distant viruses, which require a C-terminal chaperone for correct folding. The highly homologous chaperone domains are interchangeable between pre-proteins and release themselves after protein folding. Here we report the crystal structures of two intramolecular chaperone domains in either the released or the pre-cleaved form, revealing the role of the chaperone domain in the formation of a triple–β-helix fold. Tentacle-like protrusions enclose the polypeptide chains of the pre-protein during the folding process. After the assembly, a sensory mechanism for correctly folded β-helices triggers a serine-lysine catalytic dyad to autoproteolytically release the mature protein. Sequence analysis shows a conservation of the intramolecular chaperones in functionally unrelated proteins sharing β-helices as a common structural motif.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Anfinsen, C.B. Principles that govern the folding of protein chains. Science 181, 223–230 (1973).
Fenton, W.A. & Horwich, A.L. GroEL-mediated protein folding. Protein Sci. 6, 743–760 (1997).
Hartl, F.U. Molecular chaperones in cellular protein folding. Nature 381, 571–579 (1996).
Chen, Y.J. & Inouye, M. The intramolecular chaperone-mediated protein folding. Curr. Opin. Struct. Biol. 18, 765–770 (2008).
Mühlenhoff, M., Stummeyer, K., Grove, M., Sauerborn, M. & Gerardy-Schahn, R. Proteolytic processing and oligomerization of bacteriophage-derived endosialidases. J. Biol. Chem. 278, 12634–12644 (2003).
Schwarzer, D., Stummeyer, K., Gerardy-Schahn, R. & Muhlenhoff, M. Characterization of a novel intramolecular chaperone domain conserved in endosialidases and other bacteriophage tail spike and fiber proteins. J. Biol. Chem. 282, 2821–2831 (2007).
Stummeyer, K., Dickmanns, A., Muhlenhoff, M., Gerardy-Schahn, R. & Ficner, R. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 12, 90–96 (2005).
Weigele, P.R., Scanlon, E. & King, J. Homotrimeric, β-stranded viral adhesins and tail proteins. J. Bacteriol. 185, 4022–4030 (2003).
Kajava, A.V. & Steven, A.C. β-rolls, β-helices, and other β-solenoid proteins. Adv. Protein Chem. 73, 55–96 (2006).
Papanikolopoulou, K. et al. Amyloid fibril formation from sequences of a natural β-structured fibrous protein, the adenovirus fiber. J. Biol. Chem. 280, 2481–2490 (2005).
Mitraki, A., Papanikolopoulou, K. & Van Raaij, M.J. Natural triple β-stranded fibrous folds. Adv. Protein Chem. 73, 97–124 (2006).
Oinonen, C. & Rouvinen, J. Structural comparison of Ntn-hydrolases. Protein Sci. 9, 2329–2337 (2000).
Slilaty, S.N. & Vu, H.K. The role of electrostatic interactions in the mechanism of peptide bond hydrolysis by a Ser-Lys catalytic dyad. Protein Eng. 4, 919–922 (1991).
Dao-pin, S., Anderson, D.E., Baase, W.A., Dahlquist, F.W. & Matthews, B.W. Structural and thermodynamic consequences of burying a charged residue within the hydrophobic core of T4 lysozyme. Biochemistry 30, 11521–11529 (1991).
Lin, L.L. & Little, J.W. Autodigestion and RecA-dependent cleavage of Ind- mutant LexA proteins. J. Mol. Biol. 210, 439–452 (1989).
Papanikolopoulou, K., Forge, V., Goeltz, P. & Mitraki, A. Formation of highly stable chimeric trimers by fusion of an adenovirus fiber shaft fragment with the foldon domain of bacteriophage t4 fibritin. J. Biol. Chem. 279, 8991–8998 (2004).
Papanikolopoulou, K. et al. Adenovirus fibre shaft sequences fold into the native triple β-spiral fold when N-terminally fused to the bacteriophage T4 fibritin foldon trimerisation motif. J. Mol. Biol. 342, 219–227 (2004).
van Raaij, M.J., Mitraki, A., Lavigne, G. & Cusack, S. A triple β-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401, 935–938 (1999).
Barbirz, S. et al. Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol. Microbiol. 69, 303–316 (2008).
Müller, J.J. et al. An intersubunit active site between supercoiled parallel β helices in the trimeric tailspike endorhamnosidase of Shigella flexneri Phage Sf6. Structure 16, 766–775 (2008).
Steinbacher, S. et al. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267, 865–880 (1997).
Steinbacher, S. et al. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265, 383–386 (1994).
Xiang, Y. et al. Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol. Cell 34, 375–386 (2009).
Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008).
Siegert, R., Leroux, M.R., Scheufler, C., Hartl, F.U. & Moarefi, I. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell 103, 621–632 (2000).
Korndörfer, I.P., Dommel, M.K. & Skerra, A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat. Struct. Mol. Biol. 11, 1015–1020 (2004).
Stirling, P.C., Bakhoum, S.F., Feigl, A.B. & Leroux, M.R. Convergent evolution of clamp-like binding sites in diverse chaperones. Nat. Struct. Mol. Biol. 13, 865–870 (2006).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993).
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007).
Terwilliger, T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 (2004).
Terwilliger, T.C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 (2008).
Zwart, P.H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008).
Lebedev, A.A., Vagin, A.A. & Murshudov, G.N. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta Crystallogr. D Biol. Crystallogr. 64, 33–39 (2008).
McCoy, A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 (2007).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Gouet, P., Courcelle, E., Stuart, D.I. & Metoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999).
Gouet, P., Robert, X. & Courcelle, E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 (2003).
Acknowledgements
The authors would like to thank the beamline staff scientists of European Synchrotron Radiation Facility, Dr. P. Tucker from the European Molecular Biology Laboratory for outstanding support during data collection at the Deutsches Elektronen Synchrotron and D. Gloth and J. Warweg for technical assistance.
Author information
Authors and Affiliations
Contributions
E.-C.S. performed molecular cloning, protein purification and crystallization, data collection, integration and refinement, data analysis and writing of the manuscript; A.D., sequence comparison and alignments and writing of the manuscript; H.U., MS analysis; A.S., molecular cloning; M.M., K.S., D.S., feedback on the manuscript; R.G.-S., initiator of the project; R.F., data analysis, writing of the manuscript and acted as project leader.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 (PDF 5784 kb)
Rights and permissions
About this article
Cite this article
Schulz, E., Dickmanns, A., Urlaub, H. et al. Crystal structure of an intramolecular chaperone mediating triple–β-helix folding. Nat Struct Mol Biol 17, 210–215 (2010). https://doi.org/10.1038/nsmb.1746
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1746
This article is cited by
-
Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres
Nature Microbiology (2019)
-
Crystal structure of the DNA-binding domain of Myelin-gene Regulatory Factor
Scientific Reports (2017)