Abstract
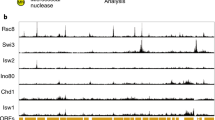
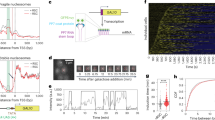
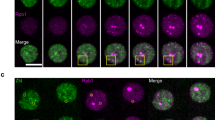
Transcription regulation in eukaryotes involves rapid recruitment and proper assembly of transcription factors at gene promoters. To determine the dynamics of the transcription machinery on DNA, we used a differential chromatin immunoprecipitation procedure coupled to whole-genome microarray detection in Saccharomyces cerevisiae. We find that TATA-binding protein (TBP) turnover is low at RNA polymerase I (Pol I) promoters. Whereas RNA polymerase III (Pol III) promoters represent an intermediate case, TBP turnover is high at RNA polymerase II (Pol II) promoters. Within these promoters, the highest turnover correlates with binding of the Spt–Ada–Gcn5 acetyltransferase complex (SAGA) coactivator, Mot1p dependence and presence of a canonical TATA box. In contrast, slow turnover Pol II promoters depend on TFIID and on the gene-specific factor, Rap1p. Together this shows that TBP turnover is regulated by protein factors rather than DNA sequence and argues that TBP turnover is an important determinant in regulating gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Albright, S.R. & Tjian, R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242, 1–13 (2000).
Thomas, M.C. & Chiang, C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 (2006).
Hernandez, N. TBP, a universal eukaryotic transcription factor? Genes Dev. 7, 1291–1308 (1993).
Basehoar, A.D., Zanton, S.J. & Pugh, B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116, 699–709 (2004).
Kim, J. & Iyer, V.R. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol. Cell. Biol. 24, 8104–8112 (2004).
Pugh, B.F. Control of gene expression through regulation of the TATA-binding protein. Gene 255, 1–14 (2000).
Steffan, J.S., Keys, D.A., Dodd, J.A. & Nomura, M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 10, 2551–2563 (1996).
Borggrefe, T., Davis, R., Bareket-Samish, A. & Kornberg, R.D. Quantitation of the RNA polymerase II transcription machinery in yeast. J. Biol. Chem. 276, 47150–47153 (2001).
Sprouse, R.O. et al. Regulation of TATA-binding protein dynamics in living yeast cells. Proc. Natl. Acad. Sci. USA 105, 13304–13308 (2008).
Hahn, S., Buratowski, S., Sharp, P.A. & Guarente, L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc. Natl. Acad. Sci. USA 86, 5718–5722 (1989).
Hoopes, B.C., LeBlanc, J.F. & Hawley, D.K. Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multi-step pathway. J. Biol. Chem. 267, 11539–11547 (1992).
van Werven, F.J. et al. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 22, 2359–2369 (2008).
Nalley, K., Johnston, S.A. & Kodadek, T. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature 442, 1054–1057 (2006).
Schermer, U.J., Korber, P. & Horz, W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell 19, 279–285 (2005).
Dion, M.F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007).
Cloutier, T.E., Librizzi, M.D., Mollah, A.K., Brenowitz, M. & Willis, I.M. Kinetic trapping of DNA by transcription factor IIIB. Proc. Natl. Acad. Sci. USA 98, 9581–9586 (2001).
Aprikian, P., Moorefield, B. & Reeder, R.H. New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol. 21, 4847–4855 (2001).
Panov, K.I., Friedrich, J.K. & Zomerdijk, J.C. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol. Cell. Biol. 21, 2641–2649 (2001).
Fan, X., Shi, H. & Lis, J.T. Distinct transcriptional responses of RNA polymerases I, II and III to aptamers that bind TBP. Nucleic Acids Res. 33, 838–845 (2005).
Harbison, C.T. et al. Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004).
Holstege, F.C. et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998).
Lee, W. et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 39, 1235–1244 (2007).
Lieb, J.D., Liu, X., Botstein, D. & Brown, P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28, 327–334 (2001).
Xu, Z. et al. Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037 (2009).
Auble, D.T. et al. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8, 1920–1934 (1994).
Andrau, J.C. et al. Mot1p is essential for TBP recruitment to selected promoters during in vivo gene activation. EMBO J. 21, 5173–5183 (2002).
Dasgupta, A., Darst, R.P., Martin, K.J., Afshari, C.A. & Auble, D.T. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. USA 99, 2666–2671 (2002).
Gilfillan, S., Stelzer, G., Piaia, E., Hofmann, M.G. & Meisterernst, M. Efficient binding of NC2.TATA-binding protein to DNA in the absence of TATA. J. Biol. Chem. 280, 6222–6230 (2005).
Schluesche, P., Stelzer, G., Piaia, E., Lamb, D.C. & Meisterernst, M. NC2 mobilizes TBP on core promoter TATA boxes. Nat. Struct. Mol. Biol. 14, 1196–1201 (2007).
Ketela, T., Brown, J.L., Stewart, R.C. & Bussey, H. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol. Gen. Genet. 259, 372–378 (1998).
Morgan, B.A. et al. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16, 1035–1044 (1997).
Sorger, P.K. Heat shock factor and the heat shock response. Cell 65, 363–366 (1991).
Garbett, K.A., Tripathi, M.K., Cencki, B., Layer, J.H. & Weil, P.A. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol. Cell. Biol. 27, 297–311 (2007).
Mencía, M., Moqtaderi, Z., Geisberg, J.V., Kuras, L. & Struhl, K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9, 823–833 (2002).
Whitehall, S.K., Kassavetis, G.A. & Geiduschek, E.P. The symmetry of the yeast U6 RNA gene's TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 9, 2974–2985 (1995).
Sprouse, R.O., Wells, M.N. & Auble, D.T. TATA-binding protein variants that bypass the requirement for Mot1 in vivo. J. Biol. Chem. 284, 4525–4535 (2009).
Darst, R.P. et al. Mot1 regulates the DNA binding activity of free TATA-binding protein in an ATP-dependent manner. J. Biol. Chem. 278, 13216–13226 (2003).
Vasiljeva, L., Kim, M., Terzi, N., Soares, L.M. & Buratowski, S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 29, 313–323 (2008).
Dudley, A.M., Rougeulle, C. & Winston, F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13, 2940–2945 (1999).
Laprade, L., Rose, D. & Winston, F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics 177, 2007–2017 (2007).
Mohibullah, N. & Hahn, S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 22, 2994–3006 (2008).
Huisinga, K.L. & Pugh, B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13, 573–585 (2004).
Venters, B.J. & Pugh, B.F. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19, 360–371 (2009).
Karpova, T.S. et al. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319, 466–469 (2008).
Yao, J., Munson, K.M., Webb, W.W. & Lis, J.T. Dynamics of heat shock factor association with native gene loci in living cells. Nature 442, 1050–1053 (2006).
Mitra, D., Parnell, E.J., Landon, J.W., Yu, Y. & Stillman, D.J. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol. Cell. Biol. 26, 4095–4110 (2006).
Whitehouse, I., Rando, O.J., Delrow, J. & Tsukiyama, T. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450, 1031–1035 (2007).
Geisberg, J.V. & Struhl, K. Cellular stress alters the transcriptional properties of promoter-bound Mot1-TBP complexes. Mol. Cell 14, 479–489 (2004).
Hsu, J.Y. et al. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 22, 2353–2358 (2008).
Huisinga, K.L. & Pugh, B.F.A. TATA binding protein regulatory network that governs transcription complex assembly. Genome Biol. 8, R46 (2007).
van Werven, F.J. & Timmers, H.T. The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 34, e33 (2006).
Longtine, M.S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (1998).
van Oevelen, C.J., van Teeffelen, H.A. & Timmers, H.T. Differential requirement of SAGA subunits for Mot1p and Taf1p recruitment in gene activation. Mol. Cell. Biol. 25, 4863–4872 (2005).
van Bakel, H. et al. Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic Acids Res. 36, e21 (2008).
Abascal, F., Carmona-Saez, P., Carazo, J.M. & Pascual-Montano, A. ChIPCodis: mining complex regulatory systems in yeast by concurrent enrichment analysis of ChIP-on-chip data. Bioinformatics 24, 1208–1209 (2008).
Acknowledgements
We are grateful to M. Groot Koerkamp, D. van Leenen and C. Ko of the University Medical Center (UMC) Utrecht and Utrecht University microarray facility for technical assistance. We are grateful to T. Weil (Vanderbilt University) for providing TBP antibodies. We also thank P. Lijnzaad and H. van Bakel for discussions and G. Spedale, P. Pijnappel and P. de Graaf for critical reading of this manuscript. This work is supported by grants (805.47.080, 825.06.033 and 700.57.302) of the Netherlands Organization for Scientific Research (NWO), the Netherlands Proteomics Centre (NPC) and the EU (Integrated Project EUTRACC, LSHG-CT-2007-037445).
Author information
Authors and Affiliations
Contributions
F.J.v.W. and H.Th.M.T. designed the experiments; F.J.W. and H.A.A.M.v.T. carried out the experiments; F.J.v.W. and H.Th.M.T. analyzed the data; F.C.P.H. provided advice on design of the experiments and data analysis; H.Th.M.T. supervised the experiments and data analysis; F.J.v.W. and H.Th.M.T. wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 1 and 2 and Supplementary Methods (PDF 1191 kb)
Rights and permissions
About this article
Cite this article
van Werven, F., van Teeffelen, H., Holstege, F. et al. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol 16, 1043–1048 (2009). https://doi.org/10.1038/nsmb.1674
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1674
This article is cited by
-
Molecular determinants underlying functional innovations of TBP and their impact on transcription initiation
Nature Communications (2020)
-
RNA synthesis is associated with multiple TBP-chromatin binding events
Scientific Reports (2017)
-
Affinity and competition for TBP are molecular determinants of gene expression noise
Nature Communications (2016)
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms
Chromosoma (2015)
-
High-resolution digital profiling of the epigenome
Nature Reviews Genetics (2014)