Abstract
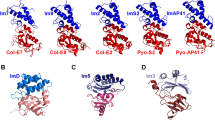
How do intricate multi-residue features such as protein-protein interfaces evolve? To address this question, we evolved a new colicin-immunity binding interaction. We started with Im9, which inhibits its cognate DNase ColE9 at 10−14 M affinity, and evolved it toward ColE7, which it inhibits promiscuously (Kd > 10−8 M). Iterative rounds of random mutagenesis and selection toward higher affinity for ColE7, and selectivity (against ColE9 inhibition), led to an ∼105-fold increase in affinity and a 108-fold increase in selectivity. Analysis of intermediates along the evolved variants revealed that changes in the binding configuration of the Im protein uncovered a latent set of interactions, thus providing the key to the rapid divergence of new Im7 variants. Overall, protein-protein interfaces seem to share the evolvability features of enzymes, that is, the exploitation of promiscuous interactions and alternative binding configurations via 'generalist' intermediates, and the key role of compensatory stabilizing mutations in facilitating the divergence of new functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Dean, A.M. & Thornton, J.W. Mechanistic approaches to the study of evolution: the functional synthesis. Nat. Rev. Genet. 8, 675–688 (2007).
Bridgham, J.T., Carroll, S.M. & Thornton, J.W. Evolution of hormone-receptor complexity by molecular exploitation. Science 312, 97–101 (2006).
Miller, S.P., Lunzer, M. & Dean, A.M. Direct demonstration of an adaptive constraint. Science 314, 458–461 (2006).
Weinreich, D.M., Delaney, N.F., Depristo, M.A. & Hartl, D.L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006).
Ortlund, E.A., Bridgham, J.T., Redinbo, M.R. & Thornton, J.W. Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317, 1544–1548 (2007).
Yokoyama, S., Tada, T., Zhang, H. & Britt, L. Elucidation of phenotypic adaptations: molecular analyses of dim-light vision proteins in vertebrates. Proc. Natl. Acad. Sci. USA 105, 13480–13485 (2008).
Aharoni, A. et al. The 'evolvability' of promiscuous protein functions. Nat. Genet. 37, 73–76 (2005).
Matsumura, I. & Ellington, A.D. In vitro evolution of β-glucuronidase into a β-galactosidase proceeds through non-specific intermediates. J. Mol. Biol. 305, 331–339 (2001).
Khersonsky, O., Roodveldt, C. & Tawfik, D.S. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10, 498–508 (2006).
Riley, M.A. Molecular mechanisms of colicin evolution. Mol. Biol. Evol. 10, 1380–1395 (1993).
Riley, M.A. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Genet. 32, 255–278 (1998).
Kleanthous, C. & Walker, D. Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem. Sci. 26, 624–631 (2001).
Reichmann, D., Rahat, O., Cohen, M., Neuvirth, H. & Schreiber, G. The molecular architecture of protein-protein binding sites. Curr. Opin. Struct. Biol. 17, 67–76 (2007).
Kühlmann, U.C., Pommer, A.J., Moore, G.R., James, R. & Kleanthous, C. Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J. Mol. Biol. 301, 1163–1178 (2000).
Bernath, K., Magdassi, S. & Tawfik, D.S. Directed evolution of protein inhibitors of DNA-nucleases by in vitro compartmentalization (IVC) and nano-droplet delivery. J. Mol. Biol. 345, 1015–1026 (2005).
Maté, M.J. & Kleanthous, C. Structure-based analysis of the metal-dependent mechanism of H-N-H endonucleases. J. Biol. Chem. 279, 34763–34769 (2004).
Li, W., Dennis, C.A., Moore, G.R., James, R. & Kleanthous, C. Protein-protein interaction specificity of Im9 for the endonuclease toxin colicin E9 defined by homologue-scanning mutagenesis. J. Biol. Chem. 272, 22253–22258 (1997).
Wallis, R. et al. In vivo and in vitro characterization of overproduced colicin E9 imunity protein. Eur. J. Biochem. 207, 687–695 (1992).
Li, W. et al. Highly discriminating protein-protein interaction specificities in the context of a conserved binding energy hotspot. J. Mol. Biol. 337, 743–759 (2004).
Keeble, A.H. & Kleanthous, C. The kinetic basis for dual recognition in colicin endonuclease-immunity protein complexes. J. Mol. Biol. 352, 656–671 (2005).
Kleanthous, C. et al. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat. Struct. Biol. 6, 243–252 (1999).
Ko, T.P., Liao, C.C., Ku, W.Y., Chak, K.F. & Yuan, H.S. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure 7, 91–102 (1999).
Lu, F.M., Yuan, H.S., Hsu, Y.C., Chang, S.J. & Chak, K.F. Hierarchical order of critical residues on the immunity-determining region of the Im7 protein which confer specific immunity to its cognate colicin. Biochem. Biophys. Res. Commun. 264, 69–75 (1999).
Wallis, R. et al. Specificity in protein-protein recognition: conserved Im9 residues are the major determinants of stability in the colicin E9 DNase-Im9 complex. Biochemistry 37, 476–485 (1998).
Kortemme, T. et al. Computational redesign of protein-protein interaction specificity. Nat. Struct. Mol. Biol. 11, 371–379 (2004).
Hiniker, A. et al. Laboratory evolution of one disulfide isomerase to resemble another. Proc. Natl. Acad. Sci. USA 104, 11670–11675 (2007).
O'Maille, P.E. et al. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4, 617–623 (2008).
Foote, J. & Milstein, C. Conformational isomerism and the diversity of antibodies. Proc. Natl. Acad. Sci. USA 91, 10370–10374 (1994).
Tokuriki, N. & Tawfik, D.S. Protein dynamism and evolvability. Science 324, 203–207 (2009).
Collins, C.H., Leadbetter, J.R. & Arnold, F.H. Dual selection enhances the signaling specificity of a variant of the quorum-sensing transcriptional activator LuxR. Nat. Biotechnol. 24, 708–712 (2006).
Ferguson, N., Capaldi, A.P., James, R., Kleanthous, C. & Radford, S.E. Rapid folding with and without populated intermediates in the homologous four-helix proteins Im7 and Im9. J. Mol. Biol. 286, 1597–1608 (1999).
Friel, C.T., Smith, D.A., Vendruscolo, M., Gsponer, J. & Radford, S.E. The mechanism of folding of Im7 reveals competition between functional and kinetic evolutionary constraints. Nat. Struct. Mol. Biol. 16, 318–324 (2009).
Tokuriki, N., Stricher, F., Serrano, L. & Tawfik, D.S. How protein stability and new functions trade off. PLOS Comput. Biol. 4, e1000002 (2008).
Bershtein, S., Goldin, K. & Tawfik, D.S. Intense neutral drifts yield robust and evolvable consensus proteins. J. Mol. Biol. 379, 1029–1044 (2008).
Wang, X., Minasov, G. & Shoichet, B.K. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 320, 85–95 (2002).
Tomatis, P.E. et al. Adaptive protein evolution grants organismal fitness by improving catalysis and flexibility. Proc. Natl. Acad. Sci. USA 105, 20605–20610 (2008).
Bloom, J.D., Labthavikul, S.T., Otey, C.R. & Arnold, F.H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA 103, 5869–5874 (2006).
Wallace, A.C., Laskowski, R.A. & Thornton, J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 (1995).
Wallis, R. et al. Protein-protein interactions in colicin E9 DNase-immunity protein complexes. 2. Cognate and noncognate interactions that span the millimolar to femtomolar affinity range. Biochemistry 34, 13751–13759 (1995).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
French, S. & Wilson, K. On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 (1978).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Acknowledgements
We gratefully acknowledge financial support by the Israel Ministry of Science and Technology, the EU training network ProSA and the Sasson and Marjorie Peress Philanthropic Fund. We are grateful to G. Schreiber, O. Cohavi and M. Harel for their help in affinity measurements. C.K. acknowledges the UK Biotechnology and Biological Sciences Research Council for funding.
Author information
Authors and Affiliations
Contributions
K.B.L. performed the selections and variant characterization; O.D. and S.A. crystallized and solved the structures; S.M. participated in the development of the emulsion selections; A.H.K. and C.K. performed stopped-flow measurements; D.S.T. designed the experiments and analyzed the data; K.B.L., O.D. and D.S.T. wrote the manuscript.
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1 and 2 (PDF 2983 kb)
Rights and permissions
About this article
Cite this article
Levin, K., Dym, O., Albeck, S. et al. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat Struct Mol Biol 16, 1049–1055 (2009). https://doi.org/10.1038/nsmb.1670
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1670
This article is cited by
-
Structural design principles for specific ultra-high affinity interactions between colicins/pyocins and immunity proteins
Scientific Reports (2021)
-
Ultrahigh specificity in a network of computationally designed protein-interaction pairs
Nature Communications (2018)
-
Computational redesign of enzymes for regio- and enantioselective hydroamination
Nature Chemical Biology (2018)
-
Analysis of Saturation Recovery Amplitudes to Characterize Conformational Exchange in Spin-Labeled Proteins
Applied Magnetic Resonance (2017)
-
Evolution acting on the same target, but at multiple levels: Proteins as the test case
Journal of Biosciences (2017)