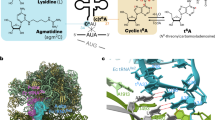
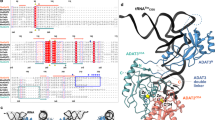
Abstract
tRNA precursors undergo a maturation process, involving nucleotide modifications and folding into the L-shaped tertiary structure. The N1-methylguanosine at position 37 (m1G37), 3′ adjacent to the anticodon, is essential for translational fidelity and efficiency. In archaea and eukaryotes, Trm5 introduces the m1G37 modification into all tRNAs bearing G37. Here we report the crystal structures of archaeal Trm5 (aTrm5) in complex with tRNALeu or tRNACys. The D2-D3 domains of aTrm5 discover and modify G37, independently of the tRNA sequences. D1 is connected to D2-D3 through a flexible linker and is designed to recognize the shape of the tRNA outer corner, as a hallmark of the completed L shape formation. This interaction by D1 lowers the Km value for tRNA, enabling the D2-D3 catalysis. Thus, we propose that aTrm5 provides the tertiary structure checkpoint in tRNA maturation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Grosjean, H. Modification and editing of RNA: historical overview and important facts to remember. in Fine-Tuning of RNA Modifications and Editing 1–22 (CPL Press, Berks, UK, 2005).
Kim, S.H. et al. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185, 435–440 (1974).
Robertus, J.D. et al. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250, 546–551 (1974).
Dirheimer, G., Keith, G., Dumas, P. & Westhof, E. Primary, secondary, and tertiary structures of tRNAs. in tRNA: Structure, Biosynthesis and Function (eds. Söll, D. & Raj Bhandary, U.L.) 93–126 (ASM, Washington DC, 1995).
Korostelev, A., Trakhanov, S., Laurberg, M. & Noller, H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126, 1065–1077 (2006).
Jühling, F. et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 (2008).
Yokoyama, S. & Nishimura, S. Modified nucleotides and codon recognition. in tRNA: Structure, Biosynthesis and Function (eds. Söll, D. & Raj Bhandary, U.L.) 207–224 (ASM, Washington DC, 1995).
Björk, G.R. Biosynthesis and function of modified nucleosides. in tRNA: Structure, Biosynthesis and Function (eds. Söll, D. & Raj Bhandary, U.L.) 165–206 (ASM, Washington DC, 1995).
Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 34, 721–733 (2006).
Grosjean, H., Edqvist, J., Stråby, K.B. & Giegé, R. Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J. Mol. Biol. 255, 67–85 (1996).
Xie, W., Liu, X. & Huang, R.H. Chemical trapping and crystal structure of a catalytic tRNA guanine transglycosylase covalent intermediate. Nat. Struct. Biol. 10, 781–788 (2003).
Losey, H.C., Ruthenburg, A.J. & Verdine, G.L. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13, 153–159 (2006).
Seif, E. & Hallberg, B.M. RNA-protein mutually induced fit: structure of E. coli isopentenyl-tRNA transferase in complex with tRNAPhe. J. Biol. Chem. 284, 6600–6604 (2009).
Hoang, C. & Ferre-D'Amare, A.R. Cocrystal structure of a tRNA ψψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107, 929–939 (2001).
Ishitani, R. et al. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 113, 383–394 (2003).
Alian, A., Lee, T.T., Griner, S.L., Stroud, R.M. & Finer-Moore, J. Structure of a TrmA-RNA complex: a consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc. Natl. Acad. Sci. USA 105, 6876–6881 (2008).
Hur, S. & Stroud, R.M. How U38, 39, and 40 of many tRNAs become the targets for pseudouridylation by TruA. Mol. Cell 26, 189–203 (2007).
Björk, G.R., Wikström, P.M. & Byström, A.S. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244, 986–989 (1989).
Hagervall, T.G., Tuohy, T.M., Atkins, J.F. & Björk, G.R. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol. 232, 756–765 (1993).
Li, J., Esberg, B., Curran, J.F. & Björk, G.R. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 271, 209–221 (1997).
Urbonavičius, J., Qian, Q., Durand, J.M., Hagervall, T.G. & Björk, G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20, 4863–4873 (2001).
Zhang, C.M., Liu, C., Slater, S. & Hou, Y.M. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNACys. Nat. Struct. Mol. Biol. 15, 507–514 (2008).
Hauenstein, S.I. & Perona, J.J. Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei. J. Biol. Chem. 283, 22007–22017 (2008).
Perret, V. et al. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature 344, 787–789 (1990).
Björk, G.R. & Kjellin-Stråby, K. Escherichia coli mutants with defects in the biosynthesis of 5-methylaminomethyl-2-thio-uridine or 1-methylguanosine in their tRNA. J. Bacteriol. 133, 508–517 (1978).
Byström, A.S. & Björk, G.R. Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA (m1G) methyltransferase in Escherichia coli K-12. Mol. Gen. Genet. 188, 440–446 (1982).
Christian, T., Evilia, C., Williams, S. & Hou, Y.M. Distinct origins of tRNA(m1G37) methyltransferase. J. Mol. Biol. 339, 707–719 (2004).
Björk, G.R. et al. A primordial tRNA modification required for the evolution of life? EMBO J. 20, 231–239 (2001).
Brulé, H., Elliott, M., Redlak, M., Zehner, Z.E. & Holmes, W.M. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43, 9243–9255 (2004).
Ahn, H.J. et al. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. EMBO J. 22, 2593–2603 (2003).
Liu, J. et al. Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 Å resolution: a novel methyltransferase fold. Proteins 53, 326–328 (2003).
Elkins, P.A. et al. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J. Mol. Biol. 333, 931–949 (2003).
Goto-Ito, S. et al. Crystal structure of archaeal tRNA(m1G37)methyltransferase aTrm5. Proteins 72, 1274–1289 (2008).
Christian, T., Evilia, C. & Hou, Y.M. Catalysis by the second class of tRNA(m1G37) methyl transferase requires a conserved proline. Biochemistry 45, 7463–7473 (2006).
Christian, T. & Hou, Y.M. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J. Mol. Biol. 373, 623–632 (2007).
Watanabe, M. et al. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. J. Biol. Chem. 272, 20146–20151 (1997).
Kuratani, M., Bessho, Y., Nishimoto, M., Grosjean, H. & Yokoyama, S. Crystal structure and mutational study of a unique SpoU family archaeal methylase that forms 2′-O-methylcytidine at position 56 of tRNA. J. Mol. Biol. 375, 1064–1075 (2008).
Grosjean, H. et al. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie 78, 488–501 (1996).
Strobel, M.C. & Abelson, J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol. Cell. Biol. 6, 2663–2673 (1986).
Grosjean, H., Droogmans, L., Giegé, R. & Uhlenbeck, O.C. Guanosine modifications in runoff transcripts of synthetic transfer RNA-Phe genes microinjected into Xenopus oocytes. Biochim. Biophys. Acta 1050, 267–273 (1990).
Hall, K.B., Sampson, J.R., Uhlenbeck, O.C. & Redfield, A.G. Structure of an unmodified tRNA molecule. Biochemistry 28, 5794–5801 (1989).
Derrick, W.B. & Horowitz, J. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 21, 4948–4953 (1993).
Perret, V. et al. Conformation in solution of yeast tRNAAsp transcripts deprived of modified nucleotides. Biochimie 72, 735–743 (1990).
Urbonavičius, J., Durand, J.M. & Björk, G.R. Three modifications in the D and T arms of tRNA influence translation in Escherichia coli and expression of virulence genes in Shigella flexneri. J. Bacteriol. 184, 5348–5357 (2002).
Li de la Sierra-Gallay, I., Mathy, N., Pellegrini, O. & Condon, C. Structure of the ubiquitous 3′ processing enzyme RNase Z bound to transfer RNA. Nat. Struct. Mol. Biol. 13, 376–377 (2006).
Minagawa, A., Ishii, R., Takaku, H., Yokoyama, S. & Nashimoto, M. The flexible arm of tRNase Z is not essential for pre-tRNA binding but affects cleavage site selection. J. Mol. Biol. 381, 289–299 (2008).
Biou, V., Yaremchuk, A., Tukalo, M. & Cusack, S. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263, 1404–1410 (1994).
Goldgur, Y. et al. The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe. Structure 5, 59–68 (1997).
Delagoutte, B., Moras, D. & Cavarelli, J. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 19, 5599–5610 (2000).
Fukai, S. et al. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103, 793–803 (2000).
Fukai, S. et al. Mechanism of molecular interactions for tRNAVal recognition by valyl-tRNA synthetase. RNA 9, 100–111 (2003).
Walter, T.S. et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14, 1617–1622 (2006).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Fukunaga, R. & Yokoyama, S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 12, 915–922 (2005).
Adams, P.D., Pannu, N.S., Read, R.J. & Brünger, A.T. Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl. Acad. Sci. USA 94, 5018–5023 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Acknowledgements
We thank M. Kimoto and I. Hirao for their assistance with the melting temperature measurements. We also thank S. Sekine for his help in collecting diffraction data and the staff of the beamlines BL41XU at SPring-8 (Harima, Japan) and NW12A at the Photon Factory (Tsukuba, Japan) for their support during data collection. This work was supported by the Targeted Proteins Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), Grant-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (JSPS) and MEXT, the 21st century COE program (Promotion of Basic Biosciences for the Understanding of Organisms' Uniqueness) and the Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms) from MEXT. S.G.-I. was supported by the 21st century COE Program, the Global COE Program and the Research Fellowship for Young Scientists from JSPS, and M.K. was supported by the Research Fellowship for Young Scientists from JSPS.
Author information
Authors and Affiliations
Contributions
S.G.-I. performed the sample preparation of aTrm5 and tRNAs, the crystal structure determinations and the biochemical assays; T.I. provided advice and helped with the experiments; M.K. prepared aTrm56 and ArcTGT; all authors discussed the results. S.G.-I., T.I. and S.Y. wrote the manuscript; S.Y. supervised the work.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Discussion (PDF 7531 kb)
Rights and permissions
About this article
Cite this article
Goto-Ito, S., Ito, T., Kuratani, M. et al. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol 16, 1109–1115 (2009). https://doi.org/10.1038/nsmb.1653
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1653
This article is cited by
-
RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning
Nature Communications (2019)
-
Backbone resonance assignment for the full length tRNA-(N1G37) methyltransferase of Pseudomonas aeruginosa
Biomolecular NMR Assignments (2019)
-
Crystal structure of the two-subunit tRNA m1A58 methyltransferase TRM6-TRM61 from Saccharomyces cerevisiae
Scientific Reports (2016)
-
Methyl transfer by substrate signaling from a knotted protein fold
Nature Structural & Molecular Biology (2016)
-
Crystal structures of the bifunctional tRNA methyltransferase Trm5a
Scientific Reports (2016)