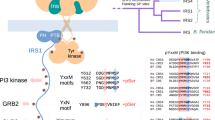
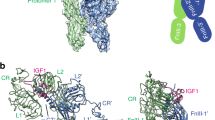
Abstract
Insulin receptor substrates 1 and 2 (IRS1 and -2) are crucial adaptor proteins in mediating the metabolic and mitogenic effects of insulin and insulin-like growth factor 1. These proteins consist of a pleckstrin homology domain, a phosphotyrosine binding domain and a C-terminal region containing numerous sites of tyrosine, serine and threonine phosphorylation. Previous yeast two-hybrid studies identified a region unique to IRS2, termed the kinase regulatory-loop binding (KRLB) region, which interacts with the tyrosine kinase domain of the insulin receptor. Here we present the crystal structure of the insulin receptor kinase in complex with a 15-residue peptide from the KRLB region. In the structure, this segment of IRS2 is bound in the kinase active site with Tyr628 positioned for phosphorylation. Although Tyr628 was phosphorylated by the insulin receptor, its catalytic turnover was poor, resulting in kinase inhibition. Our studies indicate that the KRLB region functions to limit tyrosine phosphorylation of IRS2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
White, M.F. IRS proteins and the common path to diabetes. Am. J. Physiol. Endocrinol. Metab. 283, E413–E422 (2002).
Araki, E. et al. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature 372, 186–190 (1994).
Tamemoto, H. et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature 372, 182–186 (1994).
Withers, D.J. et al. Disruption of IRS-2 causes type 2 diabetes in mice. Nature 391, 900–904 (1998).
Myers, M.G., Jr et al. IRS-1 activates phosphatidylinositol 3′-kinase by associating with src homology 2 domains of p85. Proc. Natl. Acad. Sci. USA 89, 10350–10354 (1992).
Okada, T., Kawano, Y., Sakakibara, T., Hazeki, O. & Ui, M. Essential role of phosphatidylinositol 3-kinase in insulin-induced glucose transport and antilipolysis in rat adipocytes. Studies with a selective inhibitor wortmannin. J. Biol. Chem. 269, 3568–3573 (1994).
Pirola, L., Johnston, A.M. & Van Obberghen, E. Modulation of insulin action. Diabetologia 47, 170–184 (2004).
Craparo, A., O'Neill, T.J. & Gustafson, T.A. Non-SH2 domains within insulin receptor substrate-1 and SHC mediate their phosphotyrosine-dependent interaction with the NPEY motif of the insulin-like growth factor I receptor. J. Biol. Chem. 270, 15639–15643 (1995).
Eck, M.J., Dhe-Paganon, S., Trub, T., Nolte, R.T. & Shoelson, S.E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell 85, 695–705 (1996).
Yenush, L. et al. The pleckstrin homology domain is the principal link between the insulin receptor and IRS-1. J. Biol. Chem. 271, 24300–24306 (1996).
Burks, D.J. et al. Heterologous pleckstrin homology domains do not couple IRS-1 to the insulin receptor. J. Biol. Chem. 272, 27716–27721 (1997).
Sawka-Verhelle, D., Tartare-Deckert, S., White, M.F. & Van Obberghen, E. Insulin receptor substrate-2 binds to the insulin receptor through its phosphotyrosine-binding domain and through a newly identified domain comprising amino acids 591–786. J. Biol. Chem. 271, 5980–5983 (1996).
Sawka-Verhelle, D. et al. Tyr624 and Tyr628 in insulin receptor substrate-2 mediate its association with the insulin receptor. J. Biol. Chem. 272, 16414–16420 (1997).
He, W. et al. Interaction of insulin receptor substrate-2 (IRS-2) with the insulin and insulin-like growth factor I receptors. J. Biol. Chem. 271, 11641–11645 (1996).
Hubbard, S.R. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 16, 5572–5581 (1997).
Favelyukis, S., Till, J.H., Hubbard, S.R. & Miller, W.T. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat. Struct. Biol. 8, 1058–1063 (2001).
Hubbard, S.R., Wei, L., Ellis, L. & Hendrickson, W.A. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature 372, 746–754 (1994).
Ablooglu, A.J. & Kohanski, R.A. Activation of the insulin receptor's kinase domain changes the rate-determining step of substrate phosphorylation. Biochemistry 40, 504–513 (2001).
Farooq, A., Plotnikova, O., Zeng, L. & Zhou, M.M. Phosphotyrosine binding domains of Shc and insulin receptor substrate 1 recognize the NPXpY motif in a thermodynamically distinct manner. J. Biol. Chem. 274, 6114–6121 (1999).
Barker, S.C. et al. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry 34, 14843–14851 (1995).
Miele, C. et al. Differential role of insulin receptor substrate (IRS)-1 and IRS-2 in L6 skeletal muscle cells expressing the Arg1152 → Gln insulin receptor. J. Biol. Chem. 274, 3094–3102 (1999).
Van Obberghen, E. et al. Surfing the insulin signaling web. Eur. J. Clin. Invest. 31, 966–977 (2001).
O'Neill, T.J., Craparo, A. & Gustafson, T.A. Characterization of an interaction between insulin receptor substrate 1 and the insulin receptor by using the two-hybrid system. Mol. Cell. Biol. 14, 6433–6442 (1994).
Hu, J., Liu, J., Ghirlando, R., Saltiel, A.R. & Hubbard, S.R. Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor. Mol. Cell 12, 1379–1389 (2003).
Oriente, F. et al. Insulin receptor substrate-2 phosphorylation is necessary for protein kinase C æ activation by insulin in L6hIR cells. J. Biol. Chem. 276, 37109–37119 (2001).
Schmelzle, K., Kane, S., Gridley, S., Lienhard, G.E. & White, F.M. Temporal dynamics of tyrosine phosphorylation in insulin signaling. Diabetes 55, 2171–2179 (2006).
Li, S., Covino, N.D., Stein, E.G., Till, J.H. & Hubbard, S.R. Structural and biochemical evidence for an autoinhibitory role for tyrosine 984 in the juxtamembrane region of the insulin receptor. J. Biol. Chem. 278, 26007–26014 (2003).
Acknowledgements
This work was supported by the US National Institutes of Health (NIH) grant DK052916 (to S.R.H.) and a Howard Hughes Medical Institute (HHMI) Research Training Fellowship for Medical Students (to Y.D.T.). M.F.W. is an investigator at the HHMI. We thank W. Li and W.T. Miller (Stony Brook University, New York, USA) for purified IGF1RK, and W.T. Miller for manuscript comments. Beamline X4A at the National Synchrotron Light Source, Brookhaven National Laboratory, a US Department of Energy facility, is supported by the New York Structural Biology Consortium. The New York University Protein Analysis Facility is supported by NIH Shared Instrumentation Grant S10 RR017990, National Institute of Neurological Disorders and Stroke grant P30 NS050276 and US National Cancer Institute core grant P30 CA016087 (to T.A.N.).
Author information
Authors and Affiliations
Contributions
J.W. designed and performed the crystallographic studies, the in vitro biochemical experiments and a portion of the in-cell phosphorylation experiments, and contributed to manuscript preparation. Y.D.T. designed and performed a portion of the in-cell phosphorylation experiments and contributed to manuscript preparation. C.-F.X. acquired the MS data and contributed to manuscript preparation. T.A.N. supervised the MS experiments and contributed to manuscript preparation. M.F.W. designed and supervised the in-cell biochemical experiments and contributed to manuscript preparation. S.R.H. supervised the project and was the principal manuscript author.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Methods (PDF 335 kb)
Rights and permissions
About this article
Cite this article
Wu, J., Tseng, Y., Xu, CF. et al. Structural and biochemical characterization of the KRLB region in insulin receptor substrate-2. Nat Struct Mol Biol 15, 251–258 (2008). https://doi.org/10.1038/nsmb.1388
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1388
This article is cited by
-
The combined action of the intracellular regions regulates FGFR2 kinase activity
Communications Biology (2023)
-
Circulating tumor DNA reveals mechanisms of lorlatinib resistance in patients with relapsed/refractory ALK-driven neuroblastoma
Nature Communications (2023)
-
From population to neuron: exploring common mediators for metabolic problems and mental illnesses
Molecular Psychiatry (2021)
-
A mathematical model of the impact of insulin secretion dynamics on selective hepatic insulin resistance
Nature Communications (2017)
-
Domain-dependent effects of insulin and IGF-1 receptors on signalling and gene expression
Nature Communications (2017)