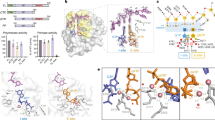
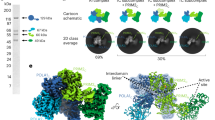
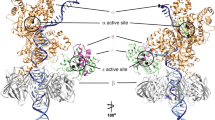
Abstract
Primases are essential RNA polymerases required for the initiation of DNA replication, lagging strand synthesis and replication restart. Many aspects of primase function remain unclear, including how the enzyme associates with a moving nucleic acid strand emanating from a helicase and orients primers for handoff to replisomal components. Using a new screening method to trap transient macromolecular interactions, we determined the structure of the Escherichia coli DnaG primase catalytic domain bound to single-stranded DNA. The structure reveals an unanticipated binding site that engages nucleic acid in two distinct configurations, indicating that it serves as a nonspecific capture and tracking locus for template DNA. Bioinformatic and biochemical analyses show that this evolutionarily constrained region enforces template polarity near the active site and is required for primase function. Together, our findings reverse previous proposals for primer–template orientation and reconcile disparate studies to re-evaluate replication fork organization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rowen, L. & Kornberg, A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J. Biol. Chem. 253, 758–764 (1978).
Kitani, T., Yoda, K., Ogawa, T. & Okazaki, T. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J. Mol. Biol. 184, 45–52 (1985).
Kornberg, A. & Baker, T. DNA Replication (Freeman, New York, 1992).
Heller, R.C. & Marians, K.J. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439, 557–562 (2006).
Frick, D.N. & Richardson, C. DNA primases. Annu. Rev. Biochem. 70, 39–80 (2001).
Kato, M., Ito, T., Wagner, G., Richardson, C. & Ellenberger, T. Modular architecture of the bacteriophage T7 primase couples RNA primer synthesis to DNA synthesis. Mol. Cell 11, 1349–1360 (2003).
Godson, G.N., Schoenich, J., Sun, W. & Mustaev, A.A. Identification of the magnesium ion binding site in the catalytic center of Escherichia coli primase by iron cleavage. Biochemistry 39, 332–339 (2000).
Aravind, L., Leipe, D. & Koonin, E. Toprim - a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26, 4205–4213 (1998).
Keck, J.L., Roche, D., Lynch, S. & Berger, J. Structure of the RNA polymerase domain of E. coli primase. Science 287, 2482–2486 (2000).
Podobnik, M., McInerney, P., O'Donnell, M. & Kuriyan, J.A. TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J. Mol. Biol. 300, 353–362 (2000).
Toth, E.A., Li, Y., Sawaya, M.R., Cheng, Y. & Ellenberger, T. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell 12, 1113–1123 (2003).
Kusakabe, T., Baradaran, K., Lee, J. & Richardson, C.C. Roles of the helicase and primase domain of the gene 4 protein of bacteriophage T7 in accessing the primase recognition site. EMBO J. 17, 1542–1552 (1998).
VanLoock, M.S., Chen, Y.J., Yu, X., Patel, S.S. & Egelman, E.H. The primase active site is on the outside of the hexameric bacteriophage T7 gene 4 helicase-primase ring. J. Mol. Biol. 311, 951–956 (2001).
Yuzhakov, A., Kelman, Z. & O'Donnell, M. Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96, 153–163 (1999).
Swart, J.R. & Griep, M.A. Primer synthesis kinetics by Escherichia coli primase on single-stranded DNA templates. Biochemistry 34, 16097–16106 (1995).
Khopde, S., Biswas, E.E. & Biswas, S.B. Affinity and sequence specificity of DNA binding and site selection for primer synthesis by Escherichia coli primase. Biochemistry 41, 14820–14830 (2002).
Corn, J.E. & Berger, J.M. FASTDXL: a generalized screen to trap disulfide-stabilized complexes for use in structural studies. Structure 15, 773–780 (2007).
Verdine, G.L. & Norman, D.P. Covalent trapping of protein-DNA complexes. Annu. Rev. Biochem. 72, 337–366 (2003).
Corn, J.E., Pease, P.J., Hura, G.L. & Berger, J.M. Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol. Cell 20, 391–401 (2005).
Lockless, S.W. & Ranganathan, R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286, 295–299 (1999).
Sun, W., Schoneich, J. & Godson, G.N. A mutant Escherichia coli primase defective in elongation of primer RNA chains. J. Bacteriol. 181, 3761–3767 (1999).
Versalovic, J. & Lupski, J.R. The Haemophilus influenzae dnaG sequence and conserved bacterial primase motifs. Gene 136, 281–286 (1993).
Brautigam, C.A. & Steitz, T.A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 8, 54–63 (1998).
Cheetham, G.M. & Steitz, T.A. Structure of a transcribing T7 RNA polymerase initiation complex. Science 286, 2305–2309 (1999).
Qimron, U., Lee, S.J., Hamdan, S.M. & Richardson, C.C. Primer initiation and extension by T7 DNA primase. EMBO J. 25, 2199–2208 (2006).
Lee, S.J. & Richardson, C.C. Interaction of adjacent primase domains within the hexameric gene 4 helicase-primase of bacteriophage T7. Proc. Natl. Acad. Sci. USA 99, 12703–12708 (2002).
Rodina, A. & Godson, G.N. Role of conserved amino acids in the catalytic activity of Escherichia coli primase. J. Bacteriol. 188, 3614–3621 (2006).
Bailey, S., Eliason, W.K. & Steitz, T.A. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318, 459–463 (2007).
MacDowell, A.A. et al. Suite of three protein crystallography beamlines with single superconducting bend magnet as the source. J. Synchrotron Radiat. 11, 447–455 (2004).
Storoni, L.C., McCoy, A.J. & Read, R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
DeLano, W.L. The PyMOL Molecular Graphics System. (DeLano Scientific, San Carlos, California, USA, 2002).
Baker, N.A., Sept, D., Joseph, S., Holst, M.J. & McCammon, J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 98, 10037–10041 (2001).
Mori, S., Abeygunawardana, C., Johnson, M.O. & van Zijl, P.C. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. B. 108, 94–98 (1995).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Shulman, A.I., Larson, C., Mangelsdorf, D.J. & Ranganathan, R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell 116, 417–429 (2004).
Acknowledgements
We thank A. Falick for assistance with mass spectrometry, C. Fromme for advice on disulfide cross-linking, J. Holton and G. Meigs of Beamline 8.3.1 for assistance with data collection, and members of the Berger laboratory for helpful discussion. This work was supported by the G. Harold and Leila Y. Mathers Foundation, and by the US National Institute of General Medical Sciences (GM071747).
Author information
Authors and Affiliations
Contributions
J.E.C. designed and performed experiments and wrote the manuscript, J.G.P. performed NMR analysis and J.M.B. supervised the experimenter, discussed results and wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figs 1–5, Methods (PDF 660 kb)
Supplementary Movie
Interpolated ssDNA tracking via the basic groove. (AVI 1830 kb)
Rights and permissions
About this article
Cite this article
Corn, J., Pelton, J. & Berger, J. Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat Struct Mol Biol 15, 163–169 (2008). https://doi.org/10.1038/nsmb.1373
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1373
This article is cited by
-
Structural Insight into the Specific DNA Template Binding to DnaG primase in Bacteria
Scientific Reports (2017)
-
Structural analysis of bacteriophage T4 DNA replication: a review in the Virology Journal series on bacteriophage T4 and its relatives
Virology Journal (2010)
-
Coordinating DNA replication by means of priming loop and differential synthesis rate
Nature (2009)
-
Understanding how the replisome works
Nature Structural & Molecular Biology (2008)