Abstract
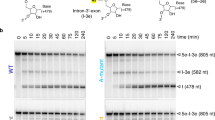
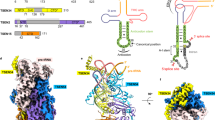
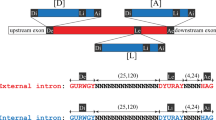
Free group II introns are infectious retroelements that can bind and insert themselves into RNA and DNA molecules via reverse splicing. Here we report the 3.4-Å crystal structure of a complex between an oligonucleotide target substrate and a group IIC intron, as well as the refined free intron structure. The structure of the complex reveals the conformation of motifs involved in exon recognition by group II introns.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Martin, W. & Koonin, E.V. Nature 440, 41–45 (2006).
Rest, J.S. & Mindell, D.P. Mol. Biol. Evol. 20, 1134–1142 (2003).
Toor, N., Hausner, G. & Zimmerly, S. RNA 7, 1142–1152 (2001).
Costa, M., Michel, F. & Westhof, E. EMBO J. 19, 5007–5018 (2000).
Pyle, A. & Lambowitz, A. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2006).
Toor, N., Keating, K.S., Taylor, S.D. & Pyle, A.M. Science 320, 77–82 (2008).
Podar, M., Perlman, P.S. & Padgett, R.A. RNA 4, 890–900 (1998).
Gordon, P.M., Fong, R. & Piccirilli, J.A. Chem. Biol. 14, 607–612 (2007).
Robart, A.R., Seo, W. & Zimmerly, S. Proc. Natl. Acad. Sci. USA 104, 6620–6625 (2007).
Acknowledgements
We thank the staff of the NE-CAT beamline 24-ID-C at the Advanced Photon Source of Argonne National Laboratory. We also thank O. Fedorova for advice and support. This work was supported by the Howard Hughes Medical Institute (HHMI) and US National Institutes of Health grant GM50313 (A.M.P.). N.T. and A.M.P. are funded by the HHMI.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2, Supplementary Table 1 and Supplementary Methods (PDF 1705 kb)
Rights and permissions
About this article
Cite this article
Toor, N., Rajashankar, K., Keating, K. et al. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol 15, 1221–1222 (2008). https://doi.org/10.1038/nsmb.1509
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1509
This article is cited by
-
Metal ions and sugar puckering balance single-molecule kinetic heterogeneity in RNA and DNA tertiary contacts
Nature Communications (2020)
-
Structure of a group II intron in complex with its reverse transcriptase
Nature Structural & Molecular Biology (2016)
-
Reverse transcriptases lend a hand in splicing catalysis
Nature Structural & Molecular Biology (2016)
-
Evolution of group II introns
Mobile DNA (2015)
-
Now on display: a gallery of group II intron structures at different stages of catalysis
Mobile DNA (2013)