Abstract
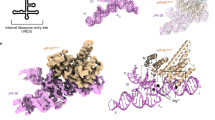
Internal ribosome entry site (IRES) RNAs initiate protein synthesis in eukaryotic cells by a noncanonical cap-independent mechanism. IRESes are critical for many pathogenic viruses, but efforts to understand their function are complicated by the diversity of IRES sequences as well as by limited high-resolution structural information. The intergenic region (IGR) IRESes of the Dicistroviridae viruses are powerful model systems to begin to understand IRES function. Here we present the crystal structure of a Dicistroviridae IGR IRES domain that interacts with the ribosome's decoding groove. We find that this RNA domain precisely mimics the transfer RNA anticodon–messenger RNA codon interaction, and its modeled orientation on the ribosome helps explain translocation without peptide bond formation. When combined with a previous structure, this work completes the first high-resolution description of an IRES RNA and provides insight into how RNAs can manipulate complex biological machines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hershey, J.W.B. & Merrick, W.C. Pathway and mechanism of initiation of protein synthesis. In Translational Control of Gene Expression (eds. Sonenberg, N., Hershey, J.W.B. & Mathews, M.B.) 33–88 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2000).
Stoneley, M. & Willis, A.E. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23, 3200–3207 (2004).
Jackson, R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 33, 1231–1241 (2005).
Schuler, M. et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 13, 1092–1096 (2006).
Spahn, C.M. et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes; the IRES functions as an RNA-based translation factor. Cell 118, 465–475 (2004).
Fukushi, S. et al. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 276, 20824–20826 (2001).
Boehringer, D., Thermann, R., Ostareck-Lederer, A., Lewis, J.D. & Stark, H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13, 1695–1706 (2005).
Spahn, C.M. et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291, 1959–1962 (2001).
Jan, E. Divergent IRES elements in invertebrates. Virus Res. 119, 16–28 (2006).
Pfingsten, J.S., Costantino, D.A. & Kieft, J.S. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J. Mol. Biol. 370, 856–869 (2007).
Costantino, D. & Kieft, J.S. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA 11, 332–343 (2005).
Nishiyama, T. et al. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 31, 2434–2442 (2003).
Jan, E. & Sarnow, P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324, 889–902 (2002).
Kanamori, Y. & Nakashima, N. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7, 266–274 (2001).
Wilson, J.E., Pestova, T.V., Hellen, C.U. & Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell 102, 511–520 (2000).
Pestova, T.V. & Hellen, C.U. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 17, 181–186 (2003).
Sasaki, J. & Nakashima, N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. USA 97, 1512–1515 (2000).
Sasaki, J. & Nakashima, N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 73, 1219–1226 (1999).
Jan, E. et al. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harb. Symp. Quant. Biol. 66, 285–292 (2001).
Thompson, S.R., Gulyas, K.D. & Sarnow, P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl. Acad. Sci. USA 98, 12972–12977 (2001).
Wilson, J.E., Powell, M.J., Hoover, S.E. & Sarnow, P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 20, 4990–4999 (2000).
Jan, E., Kinzy, T.G. & Sarnow, P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. USA 100, 15410–15415 (2003).
Pfingsten, J.S., Costantino, D.A. & Kieft, J.S. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science 314, 1450–1454 (2006).
Keel, A.Y., Rambo, R.P., Batey, R.T. & Kieft, J.S. A general strategy to solve the phase problem in RNA crystallography. Structure 15, 761–772 (2007).
Hilbers, C.W., Michiels, P.J. & Heus, H.A. New developments in structure determination of pseudoknots. Biopolymers 48, 137–153 (1998).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Cabello-Villegas, J., Tworowska, I. & Nikonowicz, E.P. Metal ion stabilization of the U-turn of the A37 N6-dimethylallyl-modified anticodon stem-loop of Escherichia coli tRNAPhe. Biochemistry 43, 55–66 (2004).
Honda, M., Brown, E.A. & Lemon, S.M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2, 955–968 (1996).
Merino, E.J., Wilkinson, K.A., Coughlan, J.L. & Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 (2005).
Wilkinson, K.A., Merino, E.J. & Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 (2006).
McGarry, K.G., Walker, S.E., Wang, H. & Fredrick, K. Destabilization of the P site codon-anticodon helix results from movement of tRNA into the P/E hybrid state within the ribosome. Mol. Cell 20, 613–622 (2005).
Moazed, D. & Noller, H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342, 142–148 (1989).
Bretscher, M.S. Translocation in protein synthesis: a hybrid structure model. Nature 218, 675–677 (1968).
Yamamoto, H., Nakashima, N., Ikeda, Y. & Uchiumi, T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J. Biol. Chem. 282, 7770–7776 (2007).
Blanchard, S.C., Kim, H.D., Gonzalez, R.L., Jr., Puglisi, J.D. & Chu, S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA 101, 12893–12898 (2004).
Munro, J.B., Altman, R.B., O'Connor, N. & Blanchard, S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell 25, 505–517 (2007).
Allen, G.S., Zavialov, A., Gursky, R., Ehrenberg, M. & Frank, J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121, 703–712 (2005).
Dorner, S., Brunelle, J.L., Sharma, D. & Green, R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat. Struct. Mol. Biol. 13, 234–241 (2006).
Olejniczak, M., Dale, T., Fahlman, R.P. & Uhlenbeck, O.C. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 12, 788–793 (2005).
Noller, H.F., Hoang, L. & Fredrick, K. The 30S ribosomal P site: a function of 16S rRNA. FEBS Lett. 579, 855–858 (2005).
Spahn, C.M. et al. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit-subunit interactions. Cell 107, 373–386 (2001).
Berk, V., Zhang, W., Pai, R.D. & Cate, J.H. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. USA 103, 15830–15834 (2006).
Valle, M. et al. Locking and unlocking of ribosomal motions. Cell 114, 123–134 (2003).
Sharma, D., Southworth, D.R. & Green, R. EF-G-independent reactivity of a pre-translocation-state ribosome complex with the aminoacyl tRNA substrate puromycin supports an intermediate (hybrid) state of tRNA binding. RNA 10, 102–113 (2004).
Kristensen, O., Laurberg, M., Liljas, A. & Selmer, M. Is tRNA binding or tRNA mimicry mandatory for translation factors? Curr. Protein Pept. Sci. 3, 133–141 (2002).
Fechter, P., Rudinger-Thirion, J., Florentz, C. & Giege, R. Novel features in the tRNA-like world of plant viral RNAs. Cell. Mol. Life Sci. 58, 1547–1561 (2001).
Moore, S.D. & Sauer, R.T. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76, 101–124 (2007).
Bessho, Y. et al. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc. Natl. Acad. Sci. USA 104, 8293–8298 (2007).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Adams, P.D. et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 (2004).
Holton, J. & Alber, T. Automated protein crystal structure determination using ELVES. Proc. Natl. Acad. Sci. USA 101, 1537–1542 (2004).
Leslie, A.G.W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newslett. Protein Crystallogr No. 26 (1992).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Terwilliger, T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 (2004).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A Foundations Crystallogr. 47, 110–119 (1991).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Kieft, J.S., Zhou, K., Jubin, R. & Doudna, J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7, 194–206 (2001).
Das, R., Laederach, A., Pearlman, S.M., Herschlag, D. & Altman, R.B. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11, 344–354 (2005).
Acknowledgements
We thank K. Bjornsen for obtaining initial crystals and for technical assistance, Q. Vicens, T. Evans, R. Batey, B. Hodges, T. Blumenthal, E. Eisenmesser, R. Zhao, M. Filbin and A. Keel for critically reading this manuscript, R. Batey for useful discussions and for providing the iridium(III) hexammine, P. Sarnow (Stanford University), E. Jan (University of British Columbia) and N. Nakashima (Japanese National Institute of Agrobiological Sciences) for plasmids, and the staffs of beam lines 4.2.2 and 12.3.1 at the Advanced Light Source. This work was supported by grants AI072187 and GM072560 from the US National Institutes of Health (J.S.K.).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Table 1, Supplementary Methods (PDF 631 kb)
Rights and permissions
About this article
Cite this article
Costantino, D., Pfingsten, J., Rambo, R. et al. tRNA–mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol 15, 57–64 (2008). https://doi.org/10.1038/nsmb1351
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1351
This article is cited by
-
A translation enhancer element from black beetle virus engages yeast eIF4G1 to drive cap-independent translation initiation
Scientific Reports (2021)
-
Viral RNA structure-based strategies to manipulate translation
Nature Reviews Microbiology (2019)
-
More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer
Cellular and Molecular Life Sciences (2017)
-
Molecular analysis of the factorless internal ribosome entry site in Cricket Paralysis virus infection
Scientific Reports (2016)
-
Initiation of translation in bacteria by a structured eukaryotic IRES RNA
Nature (2015)