Abstract
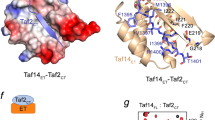
The eight twenty-one protein, ETO, is implicated in 12%–15% of acute human leukemias as part of a gene fusion with RUNX1 (also called AML1). Of the four ETO domains related to Drosophila melanogaster Nervy, only two are required to induce spontaneous myeloid leukemia upon transplantation into the mouse. One of these domains is related in sequence to TAF4, a component of TFIID. The structure of this domain, ETO-TAFH, is similar to yeast Rpb4 and to Escherichia coli σ70; it is the first TAF-related protein with structural similarity to the multisubunit RNA polymerases. Overlapping surfaces of ETO-TAFH interact with an autonomous repression domain of the nuclear receptor corepressor N-CoR and with a conserved activation domain from the E-box family of transcription factors. Thus, ETO-TAFH acts as a structural platform that can interchange negative and positive coregulatory proteins to control transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Davis, J.N., McGhee, L. & Meyers, S. The ETO (MTG8) gene family. Gene 303, 1–10 (2003).
Miyoshi, H. et al. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 88, 10431–10434 (1991).
Miyoshi, H. et al. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1–MTG8 fusion transcript. EMBO J. 12, 2715–2721 (1993).
Erickson, P. et al. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood 80, 1825–1831 (1992).
Gamou, T. et al. The partner gene of AML1 in t(16;21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood 91, 4028–4037 (1998).
Look, A.T. Oncogenic transcription factors in the human acute leukemias. Science 278, 1059–1064 (1997).
Gelmetti, V. et al. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 18, 7185–7191 (1998).
Lutterbach, B. et al. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 18, 7176–7184 (1998).
Wang, J., Hoshino, T., Redner, R.L., Kajigaya, S. & Liu, J.M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA 95, 10860–10865 (1998).
Lausen, J., Cho, S., Liu, S. & Werner, M.H. The nuclear receptor co-repressor (N-CoR) utilizes repression domains I and III for interaction and co-repression with ETO. J. Biol. Chem. 279, 49281–49288 (2004).
Minucci, S. et al. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol. Cell 5, 811–820 (2000).
Zhang, J. et al. Oligomerization of ETO is obligatory for corepressor interaction. Mol. Cell. Biol. 21, 156–163 (2001).
Liu, Y. et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer Cell 9, 249–260 (2006).
Hiebert, S.W., Lutterbach, B. & Amann, J. Role of co-repressors in transcriptional repression mediated by the t(8;21), t(16;21), t(12;21), and inv(16) fusion proteins. Curr. Opin. Hematol. 8, 197–200 (2001).
Lausen, J., Liu, S., Fliegauf, M., Lubbert, M. & Werner, M.H. ELA2 is regulated by hematopoietic transcription factors, but not repressed by AML1-ETO. Oncogene 25, 1349–1357 (2006).
Shimada, H. et al. Analysis of genes under the downstream control of the t(8;21) fusion protein AML1–MTG8: overexpression of the TIS11b (ERF-1, cMG1) gene induces myeloid cell proliferation in response to G-CSF. Blood 96, 655–663 (2000).
Andersson, A. et al. Gene expression profiling of leukemic cell lines reveals conserved molecular signatures among subtypes with specific genetic aberrations. Leukemia 19, 1042–1050 (2005).
Dunne, J. et al. siRNA-mediated AML1/MTG8 depletion affects differentiation and proliferation-associated gene expression in t(8;21)-positive cell lines and primary AML blasts. Oncogene 25, 6067–6078 (2006).
Yan, M. et al. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc. Natl. Acad. Sci. USA 101, 17186–17191 (2004).
Rhoades, K.L. et al. Analysis of the role of AML1-ETO in leukemogenesis, using an inducible transgenic mouse model. Blood 96, 2108–2115 (2000).
Yuan, Y. et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc. Natl. Acad. Sci. USA 98, 10398–10403 (2001).
Higuchi, M. et al. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell 1, 63–74 (2002).
de Guzman, C.G. et al. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol. Cell. Biol. 22, 5506–5517 (2002).
Amann, J.M. et al. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21, 6470–6483 (2001).
Zhang, J., Kalkum, M., Yamamura, S., Chait, B.T. & Roeder, R.G. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science 305, 1286–1289 (2004).
Malhotra, A., Severinova, E. & Darst, S.A. Crystal structure of a sigma 70 subunit fragment from E. coli RNA polymerase. Cell 87, 127–136 (1996).
Armache, K.J., Mitterweger, S., Meinhart, A. & Cramer, P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 280, 7131–7134 (2005).
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999).
Güntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 (2004).
Schwieters, C.D., Kuszewski, J.J., Tjandra, N. & Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003).
Clore, G.M. & Garrett, D.S. R-factor, Free R and complete cross-validation for dipolar coupling refinement of NMR structures. J. Am. Chem. Soc. 121, 9008–9012 (1999).
Spronk, C.A., Linge, J.P., Hilbers, C.W. & Vuister, G.W. Improving the quality of protein structures derived by NMR spectroscopy. J. Biomol. NMR 22, 281–289 (2002).
Plevin, M.J., Zhang, J., Guo, C., Roeder, R.G. & Ikura, M. The acute myeloid leukemia fusion protein AML1-ETO targets E proteins via a paired amphipathic helix-like TBP-associated factor homology domain. Proc. Natl. Acad. Sci. USA 103, 10242–10247 (2006).
Nomura, M., Uda-Tochio, H., Murai, K., Mori, N. & Nishimura, Y. The neural repressor NRSF/REST binds the PAH1 domain of the Sin3 corepressor by using its distinct short hydrophobic helix. J. Mol. Biol. 354, 903–915 (2005).
Swanson, K.A. et al. HBP1 and Mad1 repressors bind the Sin3 co-repressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 11, 738–746 (2004).
Spronk, C.A. et al. The Mad1-Sin3B interaction involves a novel helical fold. Nat. Struct. Biol. 7, 1100–1104 (2000).
Kolodziej, P.A., Woychik, N., Liao, S.M. & Young, R.A. RNA polymerase II subunit composition, stoichiometry, and phosphorylation. Mol. Cell. Biol. 10, 1915–1920 (1990).
Choder, M. Rpb4 and Rpb7: subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 29, 674–681 (2004).
Petermann, R. et al. Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene 17, 603–610 (1998).
Beier, D., Spohn, G., Rappuoli, R. & Scarlato, V. Functional analysis of the H. pylori principal sigma subunit of RNA polymerase reveals that the spacer region is important for efficient transcription. Mol. Microbiol. 30, 121–134 (1998).
Murre, C. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6, 1079–1086 (2005).
Massari, M.E., Jennings, P.A. & Murre, C. The AD1 transactivation domain of E2A contains a highly conserved helix which is required for its activity in both Saccharomyces cerevisiae and mammalian cells. Mol. Cell. Biol. 16, 121–129 (1996).
Dominguez, C., Boelens, R. & Bonvin, A.M.J.J. HADDOCK: a protein-protein docking approach based on biochemical and/or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003).
Zor, T., DeGuzman, R.N., Dyson, H.J. & Wright, P.E. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J. Mol. Biol. 337, 521–534 (2004).
Perissi, V. & Rosenfeld, M.G. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 6, 542–554 (2005).
Pabst, T. et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat. Med. 7, 444–445 (2001).
Tighe, J.E. & Calabi, F. Alternative, out-of-frame runt/MTG8 transcripts are encoded by the derivative (8) chromosome in the t(8;21) of acute myeloid leukemia M2. Blood 84, 2115–2121 (1994).
van de Locht, L.T., Smetsers, T.F., Wittebol, S., Raymakers, R.A. & Mensink, E.J. Molecular diversity in AML1/ETO fusion transcripts in patients with t(8;21) positive acute myeloid leukaemia. Leukemia 8, 1780–1784 (1994).
Wolford, J.K. & Prochazka, M. Structure and expression of the human MTG8/ETO gene. Gene 212, 103–109 (1998).
McNeil, S. et al. The t(8;21) chromosomal translocation in acute myelogenous leukemia modifies intranuclear targeting of the AML1/CBFalpha2 transcription factor. Proc. Natl. Acad. Sci. USA 96, 14882–14887 (1999).
Meyers, S. & Hiebert, S.W. Alterations in subnuclear trafficking of nuclear regulatory factors in acute leukemia. J. Cell. Biochem. Suppl. 35, 93–98 (2000).
Laskowski, R.A., Rullmann, J.A.C., MacArthur, M.W., Kaptein, R. & Thornton, J.M. AQUA and PROCHECK_NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996).
Hooft, R.W.W., Vriend, G., Sander, C. & Abola, E.E. Errors in protein structures. Nature 381, 272 (1996).
Acknowledgements
This work was supported in part by a fellowship from the Deutsche Forschungsgemeinschaft (LA 1389/1-1 to J.L.), by the Specialized Center of Research Grant from the Leukemia and Lymphoma Society (to M.H.W.) and by the RIKEN Structural Genomics/Proteomics Initiative, the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan. M.H.W. is a Distinguished Young Scholar of the W.M. Keck Foundation. We thank S. Hiebert (Vanderbilt University), N. Timchenko (Baylor College of Medicine), E.P. Reddy (Temple University), M.J. Klemsz (Indiana University) and S. Nimer (Memorial Sloan-Kettering Cancer Center) for providing expression and reporter vectors, T. Berg and M. Lübbert (University of Freiberg) for sharing AML1-ETO microarray data before publication and M. Punta and B. Rost for sequence analysis of the N-CoR– and E-protein–binding motifs.
Author information
Authors and Affiliations
Contributions
Y.W., S.C. and M.H.W. performed the majority of the NMR chemical shift analyses and structure calculations. Protein preparation work was carried out by C.W., Y.W. and M.H.W. S.L. and J.L. performed all of the biological assays. N.B. contributed to the NMR data analysis and computed the ETO-TAFH complex models. N.K. and S.Y. developed the KUJIRA NMR analysis toolkit.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Comparison of 2H7B and ETO-TAFH structures and their per-residue solvent accessibilities. (PDF 414 kb)
Supplementary Fig. 2
Quality assessment for structures of ETO-TAFH. (PDF 1106 kb)
Supplementary Fig. 3
1H-15N HSQC spectra of ETO-TAFH and ETO-TAFH F136Y. (PDF 683 kb)
Supplementary Fig. 4
HADDOCK modeling of the 2H7B complex with a peptide from HEB-AD1. (PDF 516 kb)
Rights and permissions
About this article
Cite this article
Wei, Y., Liu, S., Lausen, J. et al. A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription. Nat Struct Mol Biol 14, 653–661 (2007). https://doi.org/10.1038/nsmb1258
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1258
This article is cited by
-
Relationship between the structure and function of the transcriptional regulator E2A
Journal of Biological Research-Thessaloniki (2021)
-
Senataxin suppresses the antiviral transcriptional response and controls viral biogenesis
Nature Immunology (2015)
-
The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants
BMC Genomics (2010)