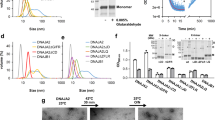
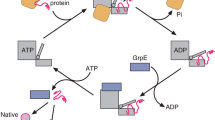
Abstract
Two members of the AAA+ superfamily, ClpB and Hsp104, collaborate with Hsp70 and Hsp40 to rescue aggregated proteins. However, the mechanisms that elicit and underlie their protein-remodeling activities remain unclear. We report that for both Hsp104 and ClpB, mixtures of ATP and ATP-γS unexpectedly unleash activation, disaggregation and unfolding activities independent of cochaperones. Mutations reveal how remodeling activities are elicited by impaired hydrolysis at individual nucleotide-binding domains. However, for some substrates, mixtures of ATP and ATP-γS abolish remodeling, whereas for others, ATP binding without hydrolysis is sufficient. Remodeling of different substrates necessitates a diverse balance of polypeptide 'holding' (which requires ATP binding but not hydrolysis) and unfolding (which requires ATP hydrolysis). We suggest that this versatility in reaction mechanism enables ClpB and Hsp104 to reactivate the entire aggregated proteome after stress and enables Hsp104 to control prion inheritance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Hanson, P.I. & Whiteheart, S.W. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 (2005).
Erzberger, J.P. & Berger, J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35, 93–114 (2006).
Hedges, S.B., Blair, J.E., Venturi, M.L. & Shoe, J.L. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 4, 2 (2004).
Sanchez, Y. & Lindquist, S.L. HSP104 required for induced thermotolerance. Science 248, 1112–1115 (1990).
Squires, C.L., Pedersen, S., Ross, B.M. & Squires, C. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 173, 4254–4262 (1991).
Sanchez, Y., Taulien, J., Borkovich, K.A. & Lindquist, S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 11, 2357–2364 (1992).
Parsell, D.A., Kowal, A.S., Singer, M.A. & Lindquist, S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372, 475–478 (1994).
Glover, J.R. & Lindquist, S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998).
Goloubinoff, P., Mogk, A., Zvi, A.P., Tomoyasu, T. & Bukau, B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 96, 13732–13737 (1999).
Zolkiewski, M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem. 274, 28083–28086 (1999).
Shorter, J. & Lindquist, S. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 6, 435–450 (2005).
Lee, S. et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115, 229–240 (2003).
Parsell, D.A., Kowal, A.S. & Lindquist, S. Saccharomyces cerevisiae Hsp104 protein. Purification and characterization of ATP-induced structural changes. J. Biol. Chem. 269, 4480–4487 (1994).
Akoev, V., Gogol, E.P., Barnett, M.E. & Zolkiewski, M. Nucleotide-induced switch in oligomerization of the AAA+ ATPase ClpB. Protein Sci. 13, 567–574 (2004).
Lum, R., Tkach, J.M., Vierling, E. & Glover, J.R. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 279, 29139–29146 (2004).
Schlieker, C. et al. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 11, 607–615 (2004).
Weibezahn, J. et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119, 653–665 (2004).
Barnett, M.E., Nagy, M., Kedzierska, S. & Zolkiewski, M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J. Biol. Chem. 280, 34940–34945 (2005).
Cashikar, A.G. et al. Defining a pathway of communication from the C-terminal peptide binding domain to the N-terminal ATPase domain in a AAA protein. Mol. Cell 9, 751–760 (2002).
Hattendorf, D.A. & Lindquist, S.L. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. EMBO J. 21, 12–21 (2002).
Mogk, A. et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 278, 17615–17624 (2003).
Schirmer, E.C., Ware, D.M., Queitsch, C., Kowal, A.S. & Lindquist, S.L. Subunit interactions influence the biochemical and biological properties of Hsp104. Proc. Natl. Acad. Sci. USA 98, 914–919 (2001).
Schlee, S., Groemping, Y., Herde, P., Seidel, R. & Reinstein, J. The chaperone function of ClpB from Thermus thermophilus depends on allosteric interactions of its two ATP-binding sites. J. Mol. Biol. 306, 889–899 (2001).
Shorter, J. & Lindquist, S. Navigating the ClpB channel to solution. Nat. Struct. Mol. Biol. 12, 4–6 (2005).
Schirmer, E.C., Homann, O.R., Kowal, A.S. & Lindquist, S. Dominant gain-of-function mutations in Hsp104p reveal crucial roles for the middle region. Mol. Biol. Cell 15, 2061–2072 (2004).
Shorter, J. & Lindquist, S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304, 1793–1797 (2004).
Shorter, J. & Lindquist, S. Destruction or potentiation of different prions catalyzed by similar hsp104 remodeling activities. Mol. Cell 23, 425–438 (2006).
Haslbeck, M., Miess, A., Stromer, T., Walter, S. & Buchner, J. Disassembling protein aggregates in the yeast cytosol. The cooperation of Hsp26 with Ssa1 and Hsp104. J. Biol. Chem. 280, 23861–23868 (2005).
Wickner, S. et al. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. USA 91, 12218–12222 (1994).
Wickner, S., Hoskins, J. & McKenney, K. Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature 350, 165–167 (1991).
Zietkiewicz, S., Lewandowska, A., Stocki, P. & Liberek, K. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J. Biol. Chem. 281, 7022–7029 (2006).
Weber-Ban, E.U., Reid, B.G., Miranker, A.D. & Horwich, A.L. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401, 90–93 (1999).
Martin, J. et al. Chaperonin-mediated protein folding at the surface of groEL through a 'molten globule'-like intermediate. Nature 352, 36–42 (1991).
Dietz, H. & Rief, M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proc. Natl. Acad. Sci. USA 101, 16192–16197 (2004).
Parsell, D.A., Sanchez, Y., Stitzel, J.D. & Lindquist, S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature 353, 270–273 (1991).
Schirmer, E.C., Queitsch, C., Kowal, A.S., Parsell, D.A. & Lindquist, S. The ATPase activity of Hsp104, effects of environmental conditions and mutations. J. Biol. Chem. 273, 15546–15552 (1998).
Barnett, M.E. & Zolkiewski, M. Site-directed mutagenesis of conserved charged amino acid residues in ClpB from Escherichia coli. Biochemistry 41, 11277–11283 (2002).
Watanabe, Y.H., Motohashi, K. & Yoshida, M. Roles of the two ATP binding sites of ClpB from Thermus thermophilus. J. Biol. Chem. 277, 5804–5809 (2002).
Weibezahn, J., Schlieker, C., Bukau, B. & Mogk, A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 278, 32608–32617 (2003).
Scheibel, T. & Lindquist, S.L. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat. Struct. Biol. 8, 958–962 (2001).
Scheibel, T. et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc. Natl. Acad. Sci. USA 100, 4527–4532 (2003).
Hersch, G.L., Burton, R.E., Bolon, D.N., Baker, T.A. & Sauer, R.T. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell 121, 1017–1027 (2005).
Whiteheart, S.W. et al. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 126, 945–954 (1994).
Wang, Q., Song, C. & Li, C.C. Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J. Struct. Biol. 146, 44–57 (2004).
Mogk, A. et al. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J. Biol. Chem. 278, 31033–31042 (2003).
Schlee, S., Beinker, P., Akhrymuk, A. & Reinstein, J. A chaperone network for the resolubilization of protein aggregates: direct interaction of ClpB and DnaK. J. Mol. Biol. 336, 275–285 (2004).
Kedzierska, S., Chesnokova, L.S., Witt, S.N. & Zolkiewski, M. Interactions within the ClpB/DnaK bi-chaperone system from Escherichia coli. Arch. Biochem. Biophys. 444, 61–65 (2005).
Zietkiewicz, S., Krzewska, J. & Liberek, K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J. Biol. Chem. 279, 44376–44383 (2004).
Martin, A., Baker, T.A. & Sauer, R.T. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature 437, 1115–1120 (2005).
Hoskins, J.R. & Wickner, S. Two peptide sequences can function cooperatively to facilitate binding and unfolding by ClpA and degradation by ClpAP. Proc. Natl. Acad. Sci. USA 103, 909–914 (2006).
Hoskins, J.R., Kim, S.Y. & Wickner, S. Substrate recognition by the ClpA chaperone component of ClpAP protease. J. Biol. Chem. 275, 35361–35367 (2000).
Shacter, E. Organic extraction of Pi with isobutanol/toluene. Anal. Biochem. 138, 416–420 (1984).
Schwede, T., Kopp, J., Guex, N. & Peitsch, M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 (2003).
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, an American Heart Association scientist development grant to J.S and NIH grants to M.Z. (GM58626) and S.L. (GM25874). We thank C. Glabe (University of California, Irvine) for the antibody to oligomer and K. Mizuuchi and K. McKenney for helpful discussions.
Author information
Authors and Affiliations
Contributions
S.M.D., J.S. and J.R.H. designed experiments, performed experiments, interpreted data and wrote the manuscript; M.Z., S.L. and S.W. designed experiments, interpreted data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Doyle, S., Shorter, J., Zolkiewski, M. et al. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol 14, 114–122 (2007). https://doi.org/10.1038/nsmb1198
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1198
This article is cited by
-
The molecular principles governing the activity and functional diversity of AAA+ proteins
Nature Reviews Molecular Cell Biology (2020)
-
Biochemical characterization of ClpB3, a chloroplastic disaggregase from Arabidopsis thaliana
Plant Molecular Biology (2020)
-
Overlapping and Specific Functions of the Hsp104 N Domain Define Its Role in Protein Disaggregation
Scientific Reports (2017)
-
Characterization of the Hsp100 disaggregase from sugarcane (SHsp101) for chaperone like activity in a yeast system
Journal of Plant Biochemistry and Biotechnology (2017)
-
Sending protein aggregates into a downward spiral
Nature Structural & Molecular Biology (2016)