Abstract
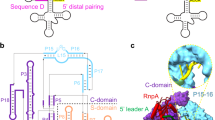
The most complex RNA pseudouridylases are H/ACA ribonucleoprotein particles, which use a guide RNA for substrate capture and four proteins (Cbf5, Nop10, Gar1 and L7Ae/NHP2) for substrate modification. Here we report the three-dimensional structure of a catalytically deficient archaeal enzyme complex (including the guide RNA and three of the four essential proteins) bound to a substrate RNA. Extensive interactions of Cbf5 with one guide-substrate helix and a guide RNA stem shape the forked guide–substrate RNA complex structure and position the substrate in proximity of the Cbf5 catalytic center. Our structural and complementary fluorescence analyses also indicate that precise placement of the target uridine at the active site requires a conformation of the guide–substrate RNA duplex that is brought about by the previously identified concurrent interaction of the guide RNA with L7Ae and a composite Cbf5-Nop10 surface, and further identify a residue that is critical in this process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hannon, G.J., Rivas, F.V., Murchison, E.P. & Steitz, J.A. The expanding universe of noncoding RNAs. Cold Spring Harb. Symp. Quant. Biol. 71, 551–564 (2006).
Matera, A.G., Terns, R.M. & Terns, M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Genet. 8, 209–220 (2007).
Huttenhofer, A. & Schattner, P. The principles of guiding by RNA: chimeric RNA-protein enzymes. Nat. Rev. Genet. 7, 475–482 (2006).
Decatur, W.A. & Fournier, M.J. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278, 695–698 (2003).
Yu, Y.T., Terns, R.M. & Terns, M.P. in Fine-tuning of RNA Functions by Modification and Editing. (ed. Grosjean, H.) 223–262 (Springer, New York, 2005).
Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20, 3617–3622 (2001).
Decatur, W.A. & Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 27, 344–351 (2002).
King, T.H., Liu, B., McCully, R.R. & Fournier, M.J. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11, 425–435 (2003).
Ni, J., Tien, A.L. & Fournier, M.J. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89, 565–573 (1997).
Ganot, P., Bortolin, M.L. & Kiss, T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89, 799–809 (1997).
Darzacq, X. et al. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 21, 2746–2756 (2002).
Eliceiri, G.L. The vertebrate E1/U17 small nucleolar ribonucleoprotein particle. J. Cell. Biochem. 98, 486–495 (2006).
Collins, K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 7, 484–494 (2006).
Omer, A.D., Ziesche, S., Decatur, W.A., Fournier, M.J. & Dennis, P.P. RNA-modifying machines in archaea. Mol. Microbiol. 48, 617–629 (2003).
Arnez, J.G. & Steitz, T.A. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33, 7560–7567 (1994).
Davis, D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 23, 5020–5026 (1995).
Newby, M.I. & Greenbaum, N.L. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7, 833–845 (2001).
Yarian, C.S. et al. Structural and functional roles of the N1- and N3-protons of psi at tRNA's position 39. Nucleic Acids Res. 27, 3543–3549 (1999).
Newby, M.I. & Greenbaum, N.L. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. USA 99, 12697–12702 (2002).
Yang, C., McPheeters, D.S. & Yu, Y.T. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J. Biol. Chem. 280, 6655–6662 (2005).
Zhao, X. & Yu, Y.T. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA 10, 681–690 (2004).
Donmez, G., Hartmuth, K. & Luhrmann, R. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA 10, 1925–1933 (2004).
Yu, Y.T., Shu, M.D. & Steitz, J.A. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17, 5783–5795 (1998).
Valadkhan, S. & Manley, J.L. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA 9, 892–904 (2003).
Yu, Y.T. The most complex pseudouridylase. Structure 14, 167–168 (2006).
Reichow, S.L., Hamma, T., Ferre-D'Amare, A.R. & Varani, G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 35, 1452–1464 (2007).
Wang, C. & Meier, U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23, 1857–1867 (2004).
Baker, D.L. et al. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 19, 1238–1248 (2005).
Charpentier, B., Muller, S. & Branlant, C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 33, 3133–3144 (2005).
Koonin, E.V. Pseudouridine synthases: four families of enzymes containing a putative uridine-binding motif also conserved in dUTPases and dCTP deaminases. Nucleic Acids Res. 24, 2411–2415 (1996).
Marrone, A., Walne, A. & Dokal, I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr. Opin. Genet. Dev. 15, 249–257 (2005).
Marrone, A. & Mason, P.J. Dyskeratosis congenita. Cell. Mol. Life Sci. 60, 507–517 (2003).
Hamma, T. & Ferre-D'Amare, A.R. Pseudouridine synthases. Chem. Biol. 13, 1125–1135 (2006).
Normand, C. et al. Analysis of the binding of the N-terminal conserved domain of yeast Cbf5p to a box H/ACA snoRNA. RNA 12, 1868–1882 (2006).
Bortolin, M.L., Ganot, P. & Kiss, T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 18, 457–469 (1999).
Manival, X. et al. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res. 34, 826–839 (2006).
Hamma, T., Reichow, S.L., Varani, G. & Ferre-D'Amare, A.R. The Cbf5–Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat. Struct. Mol. Biol. 12, 1101–1107 (2005).
Rashid, R. et al. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell 21, 249–260 (2006).
Li, L. & Ye, K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443, 302–307 (2006).
Wu, H. & Feigon, J. H/ACA small nucleolar RNA pseudouridylation pockets bind substrate RNA to form three-way junctions that position the target U for modification. Proc. Natl. Acad. Sci. USA 104, 6655–6660 (2007).
Jin, H., Loria, J.P. & Moore, P.B. Solution structure of an rRNA substrate bound to the pseudouridylation pocket of a box H/ACA snoRNA. Mol. Cell 26, 205–215 (2007).
Schattner, P. et al. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 32, 4281–4296 (2004).
Ganot, P., Caizergues-Ferrer, M. & Kiss, T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 11, 941–956 (1997).
Walne, A.J. et al. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 16, 1619–1629 (2007).
Hoang, C., Hamilton, C.S., Mueller, E.G. & Ferre-D'Amare, A.R. Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain. Protein Sci. 14, 2201–2206 (2005).
Baker, D. et al. Determination of protein-RNA interaction sites in the CBF5-H/ACA guide RNA complex by mass spectrometric protein footprinting. Biochemistry (in the press).
Hoang, C. & Ferre-D'Amare, A.R. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell 107, 929–939 (2001).
Pan, H., Agarwalla, S., Moustakas, D.T., Finer-Moore, J. & Stroud, R.M. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. USA 100, 12648–12653 (2003).
Otwinowski, Z., & Minor, W. in Processing of X-ray Diffraction Data Collected in Oscillation Mode Methods in Enzymology Vol. 276 (eds. Carter, C.W. & Sweet, R.M.) 307–326 (Academic Press, San Diego, 1997).
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D50, 760–763 (1994).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D54, 905–921 (1998).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 53, 240–255 (1997).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grant R01 GM66958-01 (H.L.) and NIH grant RO1 GM54682 (M.T. and R.T.). B. Liang is a predoctoral fellow of the American Heart Association, Florida/Puerto Rico Affiliate (0615182B). X-ray diffraction data were collected from the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions for APS beamlines are listed at http://necat.chem.cornell.edu/ and http://www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Author information
Authors and Affiliations
Contributions
B.L. designed and carried out crystallographic studies of the wild-type complex, acquired fluorescence data, and contributed to manuscript preparation; S.X. carried out crystallographic studies of the D85A mutant complex and contributed to manuscript preparation; M.P.T. and R.M.T. supplied plasmids encoding H/ACA RNP proteins and contributed to manuscript preparation; H.L. supervised the project and contributed to manuscript preparation.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1 and 2 and Supplementary Tables 1 and 2 (PDF 253 kb)
Rights and permissions
About this article
Cite this article
Liang, B., Xue, S., Terns, R. et al. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol 14, 1189–1195 (2007). https://doi.org/10.1038/nsmb1336
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1336
This article is cited by
-
RNA-dependent pseudouridylation catalyzed by box H/ACA RNPs
Frontiers in Biology (2018)