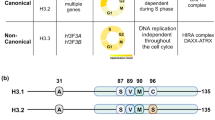
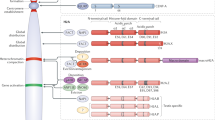
Abstract
In eukaryotes, DNA is organized into chromatin in a dynamic manner that enables it to be accessed for processes such as transcription and repair. Histones, the chief protein component of chromatin, must be assembled, replaced or exchanged to preserve or change this organization according to cellular needs. Histone chaperones are key actors during histone metabolism. Here we classify known histone chaperones and discuss how they build a network to escort histone proteins. Molecular interactions with histones and their potential specificity or redundancy are also discussed in light of chaperone structural properties. The multiplicity of histone chaperone partners, including histone modifiers, nucleosome remodelers and cell-cycle regulators, is relevant to their coordination with key cellular processes. Given the current interest in chromatin as a source of epigenetic marks, we address the potential contributions of histone chaperones to epigenetic memory and genome stability.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kornberg, R.D. Structure of chromatin. Annu. Rev. Biochem. 46, 931–954 (1977).
Oudet, P., Gross-Bellard, M. & Chambon, P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4, 281–300 (1975).
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Woodland, H.R. & Adamson, E.D. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Dev. Biol. 57, 118–135 (1977).
Loyola, A. & Almouzni, G. Histone chaperones, a supporting role in the limelight. Biochim. Biophys. Acta 1677, 3–11 (2004).
Laskey, R.A., Mills, A.D. & Morris, N.R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell 10, 237–243 (1977).
Stillman, B. Chromatin assembly during SV40 DNA replication in vitro. Cell 45, 555–565 (1986).
Smith, S. & Stillman, B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25 (1989).
Ray-Gallet, D. & Almouzni, G. DNA synthesis-dependent and -independent chromatin assembly pathways in Xenopus egg extracts. Methods Enzymol. 375, 117–131 (2004).
Quivy, J.P., Grandi, P. & Almouzni, G. Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 20, 2015–2027 (2001).
Ray-Gallet, D. et al. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9, 1091–1100 (2002).
Mello, J.A. et al. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 3, 329–334 (2002).
Ray-Gallet, D., Quivy, J.P., Sillje, H.W., Nigg, E.A. & Almouzni, G. The histone chaperone Asf1 is dispensable for direct de novo histone deposition in Xenopus egg extracts. Chromosoma 116, 487–496 (2007).
Polo, S.E. & Almouzni, G. Chromatin assembly: a basic recipe with various flavours. Curr. Opin. Genet. Dev. 16, 104–111 (2006).
Harata, M. et al. The nuclear actin-related protein of Saccharomyces cerevisiae, Act3p/Arp4, interacts with core histones. Mol. Biol. Cell 10, 2595–2605 (1999).
Luk, E. et al. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell 25, 357–368 (2007).
Loyola, A. & Almouzni, G. Marking histone H3 variants: how, when and why? Trends Biochem. Sci. 32, 425–433 (2007).
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004).
Richardson, R.T. et al. Characterization of the histone H1-binding protein, NASP, as a cell cycle-regulated somatic protein. J. Biol. Chem. 275, 30378–30386 (2000).
Shintomi, K. et al. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc. Natl. Acad. Sci. USA 102, 8210–8215 (2005).
Dutta, S. et al. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8, 841–853 (2001).
Mousson, F., Ochsenbein, F. & Mann, C. The histone chaperone Asf1 at the crossroads of chromatin and DNA checkpoint pathways. Chromosoma 116, 79–93 (2007).
Park, Y.J. & Luger, K. The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. USA 103, 1248–1253 (2006).
Belotserkovskaya, R. et al. FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003).
Angelov, D. et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 25, 1669–1679 (2006).
Daganzo, S.M. et al. Structure and function of the conserved core of histone deposition protein Asf1. Curr. Biol. 13, 2148–2158 (2003).
Muto, S. et al. Relationship between the structure of SET/TAF-Iβ/INHAT and its histone chaperone activity. Proc. Natl. Acad. Sci. USA 104, 4285–4290 (2007).
Umehara, T., Chimura, T., Ichikawa, N. & Horikoshi, M. Polyanionic stretch-deleted histone chaperone cia1/Asf1p is functional both in vivo and in vitro. Genes Cells 7, 59–73 (2002).
Regnard, C. et al. Polyglutamylation of nucleosome assembly proteins. J. Biol. Chem. 275, 15969–15976 (2000).
Namboodiri, V.M., Dutta, S., Akey, I.V., Head, J.F. & Akey, C.W. The crystal structure of Drosophila NLP-core provides insight into pentamer formation and histone binding. Structure 11, 175–186 (2003).
Mousson, F. et al. Structural basis for the interaction of Asf1 with histone H3 and its functional implications. Proc. Natl. Acad. Sci. USA 102, 5975–5980 (2005).
DeSilva, H., Lee, K. & Osley, M.A. Functional dissection of yeast Hir1p, a WD repeat-containing transcriptional corepressor. Genetics 148, 657–667 (1998).
Kaufman, P.D., Kobayashi, R., Kessler, N. & Stillman, B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell 81, 1105–1114 (1995).
Verreault, A., Kaufman, P.D., Kobayashi, R. & Stillman, B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87, 95–104 (1996).
Agez, M. et al. Structure of the histone chaperone ASF1 bound to the histone H3 C-terminal helix and functional insights. Structure 15, 191–199 (2007).
Antczak, A.J., Tsubota, T., Kaufman, P.D. & Berger, J.M. Structure of the yeast histone H3–ASF1 interaction: implications for chaperone mechanism, species-specific interactions, and epigenetics. BMC Struct. Biol. 6, 26 (2006).
English, C.M., Adkins, M.W., Carson, J.J., Churchill, M.E. & Tyler, J.K. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006).
Natsume, R. et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 (2007).
English, C.M., Maluf, N.K., Tripet, B., Churchill, M.E. & Tyler, J.K. ASF1 binds to a heterodimer of histones H3 and H4: a two-step mechanism for the assembly of the H3–H4 heterotetramer on DNA. Biochemistry 44, 13673–13682 (2005).
Schwabish, M.A. & Struhl, K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22, 415–422 (2006).
Adkins, M.W., Howar, S.R. & Tyler, J.K. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14, 657–666 (2004).
Tamburini, B.A., Carson, J.J., Adkins, M.W. & Tyler, J.K. Functional conservation and specialization among eukaryotic anti-silencing function 1 histone chaperones. Eukaryot. Cell 4, 1583–1590 (2005).
Tang, Y. et al. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat. Struct. Mol. Biol. 13, 921–929 (2006).
Sillje, H.H. & Nigg, E.A. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr. Biol. 11, 1068–1073 (2001).
Driscoll, R., Hudson, A. & Jackson, S.P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315, 649–652 (2007).
Han, J. et al. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315, 653–655 (2007).
Tsubota, T. et al. Histone H3–K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25, 703–712 (2007).
Adkins, M.W., Carson, J.J., English, C.M., Ramey, C.J. & Tyler, J.K. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J. Biol. Chem. 282, 1334–1340 (2007).
Recht, J. et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 103, 6988–6993 (2006).
Schneider, J., Bajwa, P., Johnson, F.C., Bhaumik, S.R. & Shilatifard, A. Rtt109 is required for proper H3K56 acetylation: a chromatin mark associated with the elongating RNA polymerase II. J. Biol. Chem. 281, 37270–37274 (2006).
Xhemalce, B. et al. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J. Biol. Chem. 282, 15040–15047 (2007).
Morris, S.A. et al. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 282, 7632–7640 (2007).
Lorain, S. et al. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol. Cell. Biol. 18, 5546–5556 (1998).
Sharp, J.A., Fouts, E.T., Krawitz, D.C. & Kaufman, P.D. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11, 463–473 (2001).
Zhang, R. et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30 (2005).
Tyler, J.K. et al. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21, 6574–6584 (2001).
Sanematsu, F. et al. Asf1 is required for viability and chromatin assembly during DNA replication in vertebrate cells. J. Biol. Chem. 281, 13817–13827 (2006).
Emili, A., Schieltz, D.M., Yates, J.R., III & Hartwell, L.H. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7, 13–20 (2001).
Hu, F., Alcasabas, A.A. & Elledge, S.J. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15, 1061–1066 (2001).
Gunjan, A. & Verreault, A.A. Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell 115, 537–549 (2003).
Groth, A. et al. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17, 301–311 (2005).
Polo, S.E. & Almouzni, G. Histone metabolic pathways and chromatin assembly factors as proliferation markers. Cancer Lett. 220, 1–9 (2005).
Groth, A., Rocha, W., Verreault, A. & Almouzni, G. Chromatin challenges during DNA replication and repair. Cell 128, 721–733 (2007).
Kulaeva, O.I., Gaykalova, D.A. & Studitsky, V.M. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat. Res. 618, 116–129 (2007).
Li, B., Carey, M. & Workman, J.L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Workman, J.L. Nucleosome displacement in transcription. Genes Dev. 20, 2009–2017 (2006).
Krogan, N.J. et al. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22, 6979–6992 (2002).
Bhaumik, S.R., Smith, E. & Shilatifard, A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14, 1008–1016 (2007).
Masumoto, H., Hawke, D., Kobayashi, R. & Verreault, A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 (2005).
Xu, F., Zhang, K. & Grunstein, M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121, 375–385 (2005).
Han, J., Zhou, H., Li, Z., Xu, R.M. & Zhang, Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J. Biol. Chem. 282, 14158–14164 (2007).
Duina, A.A. et al. Evidence that the localization of the elongation factor Spt16 across transcribed genes is dependent upon histone H3 integrity in Saccharomyces cerevisiae. Genetics 177, 101–112 (2007).
Adkins, M.W. & Tyler, J.K. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21, 405–416 (2006).
Kaplan, C.D., Laprade, L. & Winston, F. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301, 1096–1099 (2003).
Cairns, B.R. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 14, 989–996 (2007).
Saha, A., Wittmeyer, J. & Cairns, B.R. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7, 437–447 (2006).
Raisner, R.M. et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123, 233–248 (2005).
Thiriet, C. & Hayes, J.J. Replication-independent core histone dynamics at transcriptionally active loci in vivo. Genes Dev. 19, 677–682 (2005).
Levchenko, V., Jackson, B. & Jackson, V. Histone release during transcription: displacement of the two H2A–H2B dimers in the nucleosome is dependent on different levels of transcription-induced positive stress. Biochemistry 44, 5357–5372 (2005).
Chen, H., Li, B. & Workman, J.L. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 13, 380–390 (1994).
Walter, P.P., Owen-Hughes, T.A., Cote, J. & Workman, J.L. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell. Biol. 15, 6178–6187 (1995).
Orphanides, G., LeRoy, G., Chang, C.H., Luse, D.S. & Reinberg, D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92, 105–116 (1998).
Orphanides, G., Wu, W.H., Lane, W.S., Hampsey, M. & Reinberg, D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400, 284–288 (1999).
Ito, T., Ikehara, T., Nakagawa, T., Kraus, W.L. & Muramatsu, M. p300-mediated acetylation facilitates the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14, 1899–1907 (2000).
Swaminathan, V., Kishore, A.H., Febitha, K.K. & Kundu, T.K. Human histone chaperone nucleophosmin enhances acetylation-dependent chromatin transcription. Mol. Cell. Biol. 25, 7534–7545 (2005).
Zlatanova, J., Seebart, C. & Tomschik, M. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 21, 1294–1310 (2007).
Korber, P. et al. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J. Biol. Chem. 281, 5539–5545 (2006).
Owen-Hughes, T. & Workman, J.L. Remodeling the chromatin structure of a nucleosome array by transcription factor-targeted trans-displacement of histones. EMBO J. 15, 4702–4712 (1996).
Jamai, A., Imoberdorf, R.M. & Strubin, M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25, 345–355 (2007).
Dion, M.F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007).
Formosa, T. et al. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162, 1557–1571 (2002).
Schermer, U.J., Korber, P. & Horz, W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell 19, 279–285 (2005).
Endoh, M. et al. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol. Cell. Biol. 24, 3324–3336 (2004).
Chimura, T., Kuzuhara, T. & Horikoshi, M. Identification and characterization of CIA/ASF1 as an interactor of bromodomains associated with TFIID. Proc. Natl. Acad. Sci. USA 99, 9334–9339 (2002).
Simic, R. et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22, 1846–1856 (2003).
Konev, A.Y. et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317, 1087–1090 (2007).
Brickner, D.G. et al. H2A.Z-mediatedlocalization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5, e81 (2007).
Leno, G.H., Mills, A.D., Philpott, A. & Laskey, R.A. Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization. J. Biol. Chem. 271, 7253–7256 (1996).
Downs, J.A., Nussenzweig, M.C. & Nussenzweig, A. Chromatin dynamics and the preservation of genetic information. Nature 447, 951–958 (2007).
Bao, Y. & Shen, X. Chromatin remodeling in DNA double-strand break repair. Curr. Opin. Genet. Dev. 17, 126–131 (2007).
Altaf, M., Saksouk, N. & Cote, J. Histone modifications in response to DNA damage. Mutat. Res. 618, 81–90 (2007).
Osley, M.A., Tsukuda, T. & Nickoloff, J.A. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res. 618, 65–80 (2007).
Rodrigue, A. et al. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 25, 222–231 (2006).
Soutoglou, E. et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9, 675–682 (2007).
Berkovich, E., Monnat, R.J. Jr. & Kastan, M.B. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9, 683–690 (2007).
Rogakou, E.P., Boon, C., Redon, C. & Bonner, W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Fillingham, J., Keogh, M.C. & Krogan, N.J. GammaH2AX and its role in DNA double-strand break repair. Biochem. Cell Biol. 84, 568–577 (2006).
Bewersdorf, J., Bennett, B.T. & Knight, K.L. H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy. Proc. Natl. Acad. Sci. USA 103, 18137–18142 (2006).
Kent, N.A., Chambers, A.L. & Downs, J.A. Dual chromatin-remodelling roles for RSC during DNA double-strand break induction and repair at the yeast MAT locus. J. Biol. Chem. 282, 27693–27701 (2007).
Kim, J.A., Kruhlak, M., Dotiwala, F., Nussenzweig, A. & Haber, J.E. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J. Cell Biol. 178, 209–218 (2007).
Downs, J.A. et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990 (2004).
Morrison, A.J. et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119, 767–775 (2004).
Shen, X., Ranallo, R., Choi, E. & Wu, C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12, 147–155 (2003).
van Attikum, H., Fritsch, O., Hohn, B. & Gasser, S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788 (2004).
Qin, S. & Parthun, M.R. Recruitment of the type B histone acetyltransferase Hat1p to chromatin is linked to DNA double-strand breaks. Mol. Cell. Biol. 26, 3649–3658 (2006).
Parthun, M.R., Widom, J. & Gottschling, D.E. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87, 85–94 (1996).
Tamburini, B.A. & Tyler, J.K. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 25, 4903–4913 (2005).
Chowdhury, D. et al. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20, 801–809 (2005).
Keogh, M.C. et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439, 497–501 (2006).
Cheung, W.L. et al. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15, 656–660 (2005).
Papamichos-Chronakis, M., Krebs, J.E. & Peterson, C.L. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20, 2437–2449 (2006).
Squatrito, M., Gorrini, C. & Amati, B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16, 433–442 (2006).
Shim, E.Y., Ma, J.L., Oum, J.H., Yanez, Y. & Lee, S.E. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 25, 3934–3944 (2005).
Chai, B., Huang, J., Cairns, B.R. & Laurent, B.C. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19, 1656–1661 (2005).
Polo, S.E., Roche, D. & Almouzni, G. New histone incorporation marks sites of UV repair in human cells. Cell 127, 481–493 (2006).
Tyler, J.K. et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 (1999).
Kuzuhara, T. & Horikoshi, M. A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat. Struct. Mol. Biol. 11, 275–283 (2004).
Kleinschmidt, J.A., Fortkamp, E., Krohne, G., Zentgraf, H. & Franke, W.W. Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J. Biol. Chem. 260, 1166–1176 (1985).
Bortvin, A. & Winston, F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272, 1473–1476 (1996).
Huang, S. et al. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc. Natl. Acad. Sci. USA 102, 13410–13415 (2005).
Laskey, R.A., Honda, B.M., Mills, A.D. & Finch, J.T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275, 416–420 (1978).
Okuwaki, M., Matsumoto, K., Tsujimoto, M. & Nagata, K. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 506, 272–276 (2001).
Rougeulle, C. & Avner, P. Cloning and characterization of a murine brain specific gene Bpx and its human homologue lying within the Xic candidate region. Hum. Mol. Genet. 5, 41–49 (1996).
Okuwaki, M. & Nagata, K. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J. Biol. Chem. 273, 34511–34518 (1998).
Wang, G.S. et al. Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron 42, 113–128 (2004).
Selth, L. & Svejstrup, J.Q. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 282, 12358–12362 (2007).
Ai, X. & Parthun, M.R. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14, 195–205 (2004).
Loyola, A., LeRoy, G., Wang, Y.H. & Reinberg, D. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 15, 2837–2851 (2001).
Peterson, C.L., Zhao, Y. & Chait, B.T. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J. Biol. Chem. 273, 23641–23644 (1998).
Ito, T., Bulger, M., Pazin, M.J., Kobayashi, R. & Kadonaga, J.T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145–155 (1997).
Acknowledgements
We thank E. Dunleavy, D. Ray-Gallet and A. Gérard for their input. The authors are supported by Cancéropôle Ile-de-France (L.D.K. and G.A.); Université Paris 6 (A.C.); the Ligue Nationale contre le Cancer (Equipe labellisée la Ligue); the Commissariat à l'Energie Atomique (LRC no. 26), European Contract RTN (HPRN-CT-2002-00238), Network of Excellence Epigenome (LSHG-CT-2004-503433), Action Concertée Interface Physique-Chimie-Biologie (04393), the Agence Nationale de la Recherche (NT05-4-422267 and PCV06-142302), and Programme Incitatif et Collaboratif (PIC) Rétinoblastome (G.A.); and US National Institutes of Health grants GM61766 and GM20056 (J.E.H.). J.E.H. received a Mayent-Rotschild sabbatical fellowship from the Institut Curie. We apologize to authors whose work has not been cited because of limited space.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
De Koning, L., Corpet, A., Haber, J. et al. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol 14, 997–1007 (2007). https://doi.org/10.1038/nsmb1318
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1318
This article is cited by
-
Structural comparisons reveal diverse binding modes between nucleosome assembly proteins and histones
Epigenetics & Chromatin (2022)
-
NPM2 in malignant peritoneal mesothelioma: from basic tumor biology to clinical medicine
World Journal of Surgical Oncology (2022)
-
The histone chaperone Nrp1 is required for chromatin stability and nuclear division in Tetrahymena thermophila
Epigenetics & Chromatin (2021)
-
Nucleus-specific linker histones Hho1 and Mlh1 form distinct protein interactions during growth, starvation and development in Tetrahymena thermophila
Scientific Reports (2020)
-
OsChz1 acts as a histone chaperone in modulating chromatin organization and genome function in rice
Nature Communications (2020)