Abstract
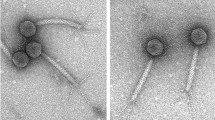
Lactococcus lactis is a Gram-positive bacterium used extensively by the dairy industry for the manufacture of fermented milk products. The double-stranded DNA bacteriophage p2 infects specific L. lactis strains using a receptor-binding protein (RBP) located at the tip of its noncontractile tail. We have solved the crystal structure of phage p2 RBP, a homotrimeric protein composed of three domains: the shoulders, a β-sandwich attached to the phage; the neck, an interlaced β-prism; and the receptor-recognition head, a seven-stranded β-barrel. We used the complex of RBP with a neutralizing llama VHH domain to identify the receptor-binding site. Structural similarity between the recognition-head domain of phage p2 and those of adenoviruses and reoviruses, which invade mammalian cells, suggests that these viruses, despite evolutionary distant targets, lack of sequence similarity and the different chemical nature of their genomes (DNA versus RNA), might have a common ancestral gene.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Moineau, S., Tremblay, D. & Labrie, S. Phages of lactic acid bacteria: from genomics to industrial applications. ASM News 68, 388–393 (2002).
Labrie, S. & Moineau, S. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296, 308–320 (2002).
Jarvis, A.W. et al. Species and type phages of lactococcal bacteriophages. Intervirology 32, 2–9 (1991).
Valyasevi, R., Sandine, W.E. & Geller, B.L. The bacteriophage kh receptor of Lactococcus lactis subsp. cremoris KH is the rhamnose of the extracellular wall polysaccharide. Appl. Environ. Microbiol. 56, 1882–1889 (1990).
Dupont, K., Vogensen, F.K., Neve, H., Bresciani, J. & Josephsen, J. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ. Microbiol. 70, 5818–5824 (2004).
Duplessis, M. & Moineau, S. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41, 325–336 (2001).
Schafer, A., Geis, A., Neve, H. & Teuber, M. Bacteriophage receptors of Lactococcus lactis subsp. diacetylactis F7/2 and Lactococcus lactis subsp. cremoris Wg2–1. FEMS Microbiol. Lett. 62, 69–73 (1991).
Muyldermans, S., Cambillau, C. & Wyns, L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem. Sci. 26, 230–235 (2001).
Ledeboer, A.M. et al. Preventing phage lysis of Lactococcus lactis in cheese production using a neutralizing heavy-chain antibody fragment from llama. J. Dairy Sci. 85, 1376–1382 (2002).
De Haard, H.J. et al. Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection. J. Bacteriol. 187, 4531–4541 (2005).
Chappell, J.D., Prota, A.E., Dermody, T.S. & Stehle, T. Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21, 1–11 (2002).
Burmeister, W.P., Guilligay, D., Cusack, S., Wadell, G. & Arnberg, N. Crystal structure of species D adenovirus fiber knobs and their sialic acid binding sites. J. Virol. 78, 7727–7736 (2004).
van Raaij, M.J., Mitraki, A., Lavigne, G. & Cusack, S. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401, 935–938 (1999).
van Raaij, M.J., Schoehn, G., Burda, M.R. & Miller, S. Crystal structure of a heat and protease-stable part of the bacteriophage T4 short tail fibre. J. Mol. Biol. 314, 1137–1146 (2001).
Kanamaru, S. et al. Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 (2002).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Thomassen, E. et al. The structure of the receptor-binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal-binding fold. J. Mol. Biol. 331, 361–373 (2003).
Xu, L., Benson, S.D., Butcher, S.J., Bamford, D.H. & Burnett, R.M. The receptor binding protein P2 of PRD1, a virus targeting antibiotic-resistant bacteria, has a novel fold suggesting multiple functions. Structure (Camb). 11, 309–322 (2003).
Fass, D. et al. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science 277, 1662–1666 (1997).
Gibbons, D.L. et al. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427, 320–325 (2004).
Modis, Y., Ogata, S., Clements, D. & Harrison, S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 (2004).
Dormitzer, P.R., Sun, Z.Y., Wagner, G. & Harrison, S.C. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21, 885–897 (2002).
Desmyter, A. et al. Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology. J. Biol. Chem. 277, 23645–23650 (2002).
Bourne, Y. et al. Structures of a legume lectin complexed with the human lactotransferrin N2 fragment, and with an isolated biantennary glycopeptide: role of the fucose moiety. Structure 2, 209–219 (1994).
Bourne, Y. et al. Three-dimensional structures of complexes of Lathyrus ochrus isolectin I with glucose and mannose: fine specificity of the monosaccharide-binding site. Proteins 8, 365–376 (1990).
Chandry, P.S., Moore, S.C., Boyce, J.D., Davidson, B.E. & Hillier, A.J. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol. Microbiol. 26, 49–64 (1997).
Crutz-Le Coq, A.M., Cesselin, B., Commissaire, J. & Anba, J. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiol. 148, 985–1001 (2002).
George, D.G., Yeh, L.S. & Barker, W.C. Unexpected relationships between bacteriophage lambda hypothetical proteins and bacteriophage T4 tail-fiber proteins. Biochem. Biophys. Res. Commun. 115, 1061–1068 (1983).
Tetart, F. et al. Phylogeny of the major head and tail genes of the wide-ranging T4-type bacteriophages. J. Bacteriol. 183, 358–366 (2001).
Hendrix, R.W. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61, 471–480 (2002).
Bamford, D.H. Do viruses form lineages across different domains of life? Res. Microbiol. 154, 231–236 (2003).
Benson, S.D., Bamford, J.K., Bamford, D.H. & Burnett, R.M. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16, 673–685 (2004).
Rice, G. et al. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc. Natl. Acad. Sci. USA 101, 7716–7720 (2004).
Gibbons, D.L. et al. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell 114, 573–583 (2003).
Benson, S.D., Bamford, J.K., Bamford, D.H. & Burnett, R.M. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98, 825–833 (1999).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–766 (1994).
Leslie, A.G. Integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 55, 1696–1702 (1999).
Terwilliger, T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 (2004).
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Morris, R.J., Perrakis, A. & Lamzin, V.S. ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 (2003).
Murshudov, G., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Roussel, A. & Cambillau, C. The TURBO-FRODO graphics package. in Silicon Graphics Geometry Partners Directory 86 (Silicon Graphics, Mountain View, USA, 1991).
Laskowski, R., MacArthur, M., Moss, D. & Thornton, J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 91–97 (1993).
Acknowledgements
This work was supported by the Marseille-Nice Genopole, by the European Union's Structural Proteomics in Europe program (fifth PCRDT, QLG2-CT-2002-00988) and by a grant from the Natural Sciences and Engineering Research Council of Canada. C. Huyghe is greatly acknowledged for protein production and D. Tremblay (Laval University, Quebec, Canada) for phage p2 DNA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Top view of the p2 rbp head–VHH5 complex. (PDF 990 kb)
Supplementary Fig. 2
Electron density map of residues 230–234. (PDF 806 kb)
Supplementary Table 1
Interactions of the receptor binding domain with VHH mono5 (PDF 53 kb)
Rights and permissions
About this article
Cite this article
Spinelli, S., Desmyter, A., Verrips, C. et al. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat Struct Mol Biol 13, 85–89 (2006). https://doi.org/10.1038/nsmb1029
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1029
This article is cited by
-
Phage diversity, genomics and phylogeny
Nature Reviews Microbiology (2020)
-
Molecular, physiological and phylogenetic traits of Lactococcus 936-type phages from distinct dairy environments
Scientific Reports (2018)
-
Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex
Nature Microbiology (2017)
-
Structure of the host-recognition device of Staphylococcus aureus phage ϕ11
Scientific Reports (2016)
-
Crystal structure of the fibre head domain of bovine adenovirus 4, a ruminant atadenovirus
Virology Journal (2015)