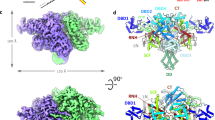
Abstract
Mobile elements and their inactive remnants account for large proportions of most eukaryotic genomes, where they have had central roles in genome evolution. Over 50 years ago, McClintock reported a form of stress-induced genome instability in maize in which discrete DNA segments move between chromosomal locations. Our current mechanistic understanding of enzymes catalyzing transposition is largely limited to prokaryotic transposases. The Hermes transposon from the housefly is part of the eukaryotic hAT superfamily that includes hobo from Drosophila, McClintock's maize Activator and Tam3 from snapdragon. We report here the three-dimensional structure of a functionally active form of the transposase from Hermes at 2.1-Å resolution. The Hermes protein has some structural features of prokaryotic transposases, including a domain with a retroviral integrase fold. However, this domain is disrupted by the insertion of an additional domain. Finally, transposition is observed only when Hermes assembles into a hexamer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Kazazian, H.H. Mobile elements: drivers of genome evolution. Science 303, 1626–1632 (2004).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Ostertag, E.M. & Kazazian, H.H., Jr. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35, 501–538 (2001).
Plasterk, R.H.A. & van Luenen, H.G.A.M. In Mobile DNA II (ed. Craig, N.L., Craigie, R., Gellert, M. & Lambowitz, A.M.) 519–532 (ASM Press, Washington, DC, USA, 2002).
Calvi, B.R., Hong, T.J., Findley, S.D. & Gelbart, W.M. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator, and Tam3. Cell 66, 465–471 (1991).
McClintock, B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 16, 13–47 (1951).
Warren, W.D., Atkinson, P.W. & O'Brochta, D.A. The Hermes transposable element from the house fly, Musca domestica, is a short inverted repeat-type element of the hobo, Ac, and Tam3 (hAT) element family. Genet. Res. 64, 87–97 (1994).
Zhou, L. et al. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature 432, 995–1001 (2004).
Roth, D.B., Menetski, J.P., Nakajima, P.B., Bosma, M.J. & Gellert, M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell 70, 983–991 (1992).
McBlane, J.F. et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83, 387–395 (1995).
Perez, Z.N., Musingarimi, P., Craig, N.L., Dyda, F. & Hickman, A.B. Purification, crystallization, and preliminary crystallographic analysis of the Hermes transposase. Acta Crystallogr. F61, 587–590 (2005).
Dyda, F. et al. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266, 1981–1986 (1994).
Curcio, M.J. & Derbyshire, K.M. The outs and ins of transposition: from Mu to kangaroo. Nat. Rev. Mol. Cell Biol. 4, 865–877 (2003).
Michel, K. & Atkinson, P.W. Nuclear localization of the Hermes transposase depends on basic amino acid residues at the N-terminus of the protein. J. Cell. Biochem. 89, 778–790 (2003).
Aravind, L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem. Sci. 25, 421–423 (2000).
Holm, L. & Sander, C. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 25, 231–234 (1997).
Davies, D.R., Goryshin, I.Y., Reznikoff, W.S. & Rayment, I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289, 77–85 (2000).
Ason, B. & Reznikoff, W.S. Mutational analysis of the base flipping event found in Tn5 transposition. J. Biol. Chem. 277, 11284–11291 (2002).
Feldmar, S. & Kunze, R. The ORFa protein, the putative transposase of maize transposable element Ac, has a basic DNA binding domain. EMBO J. 10, 4003–4010 (1991).
Marchler-Bauer, A. et al. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33, D192–D196 (2005).
Thompson, C.B. New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 3, 531–539 (1995).
van Gent, D.C., Mizuuchi, K. & Gellert, M. Similarities between initiation of V(D)J recombination and retroviral integration. Science 271, 1592–1594 (1996).
Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999).
Landree, M.A., Wibbenmeyer, J.A. & Roth, D.B. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 13, 3059–3069 (1999).
Kim, D.R., Dai, Y., Mundy, C.L., Yang, W. & Oettinger, M.A. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13, 3070–3080 (1999).
Huye, L.E., Purugganan, M.M., Jiang, M.M. & Roth, D.B. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell. Biol. 22, 3460–3473 (2002).
De, P. & Rodgers, K.K. Putting the pieces together: identification and characterization of structural domains in the V(D)J recombination protein RAG1. Immunol. Rev. 200, 70–82 (2004).
Larsen, T.A., Olson, A.J. & Goodsell, D.S. Morphology of protein-protein interfaces. Structure 6, 421–427 (1998).
Michel, K., O'Brochta, D.A. & Atkinson, P.W. The C-terminus of the Hermes transposase contains a protein multimerization domain. Insect Biochem. Mol. Biol. 33, 959–970 (2003).
Yuan, J.F., Beniac, D.R., Chaconas, G. & Ottensmeyer, F.P. 3D reconstruction of the Mu transposase and the Type 1 transpososome: a structural framework for Mu DNA transposition. Genes Dev. 19, 840–852 (2005).
Namgoong, S.Y. & Harshey, R.M. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 17, 3775–3785 (1998).
Williams, T.L., Jackson, E.L., Carritte, A. & Baker, T.A. Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev. 13, 2725–2737 (1999).
Kennedy, A.K., Haniford, D.B. & Mizuuchi, K. Single active site catalysis of the successive phosphoryl transfer steps by DNA transposases: Insights from phosphorothioate stereoselectivity. Cell 101, 295–305 (2000).
Smit, A.F.A. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9, 657–663 (1999).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Sheldrick, G.M. SHELX: applications to macromolecules. in Direct Methods for Solving Macromolecular Structures (S. Fortier, ed.) 401–411 (Kluwer, Dordrecht, The Netherlands, 1998).
Furey, W. & Swaminathan, S. PHASES-95: a program package for processing and analyzing diffraction data from macromolecules. Methods Enzymol. 277, 590–620 (1997).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47, 110–119 (1991).
Brünger, A.T. et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D54, 905–921 (1998).
Christopher, J.A. SPOCK The Structural Properties Observation and Calculation Kit (Program Manual) (The Center for Macromolecular Design, Texas A&M University, College Station, Texas, USA, 1998).
Persistence of Vision Pty. Ltd. Persistence of Vision Raytracer (Persistence of Vision Pty. Ltd., Williamstown, Victoria, Australia, 2004).
DeLano, W.L. The PyMol Molecular Graphics System (DeLano Scientific, San Carlos, California, USA, 2002).
Acknowledgements
We thank M. Gellert, S. Vasudevan and D. Ronning for helpful discussions and comments on the manuscript. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38. This study used the high-performance computational capabilities of the Helix Systems at the National Institutes of Health, Bethesda, MD (http://helix.nih.gov). N.L.C. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Electron density maps of Hermes 70–612. (PDF 11787 kb)
Supplementary Fig. 2
DNA binding by Hermes, assessed by co-elution during size exclusion chromatography, requires the 79–150 domain. (PDF 1206 kb)
Supplementary Fig. 3
Sedimentation equilibrium profiles for heterotetrameric (D1) and hexameric (H) Hermes 79–612. (PDF 2439 kb)
Rights and permissions
About this article
Cite this article
Hickman, A., Perez, Z., Zhou, L. et al. Molecular architecture of a eukaryotic DNA transposase. Nat Struct Mol Biol 12, 715–721 (2005). https://doi.org/10.1038/nsmb970
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb970
This article is cited by
-
Zinc-finger BED domains drive the formation of the active Hermes transpososome by asymmetric DNA binding
Nature Communications (2023)
-
Structure and Function of N-Terminal Zinc Finger Domain of SARS-CoV-2 NSP2
Virologica Sinica (2021)
-
DNA melting initiates the RAG catalytic pathway
Nature Structural & Molecular Biology (2018)
-
The N-terminal zinc finger domain of Tgf2 transposase contributes to DNA binding and to transposition activity
Scientific Reports (2016)
-
Biochemical Characterization of Kat1: a Domesticated hAT-Transposase that Induces DNA Hairpin Formation and MAT-Switching
Scientific Reports (2016)