Abstract
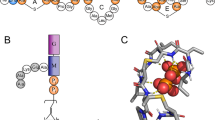
The emerging antibiotics-resistance problem has underlined the urgent need for novel antimicrobial agents. Lantibiotics (lanthionine-containing antibiotics) are promising candidates to alleviate this problem. Nisin, a member of this family, has a unique pore-forming activity against bacteria. It binds to lipid II, the essential precursor of cell wall synthesis. As a result, the membrane permeabilization activity of nisin is increased by three orders of magnitude. Here we report the solution structure of the complex of nisin and lipid II. The structure shows a novel lipid II–binding motif in which the pyrophosphate moiety of lipid II is primarily coordinated by the N-terminal backbone amides of nisin via intermolecular hydrogen bonds. This cage structure provides a rationale for the conservation of the lanthionine rings among several lipid II–binding lantibiotics. The structure of the pyrophosphate cage offers a template for structure-based design of novel antibiotics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Koch, A.L. Bacterial wall as target for attack: Past, present, and future research. Clin. Microbiol. Rev. 16, 673–687 (2003).
van Heijenoort, J. Biosynthesis of the bacterial peptidoglycan unit. In Bacterial Cell Wall Vol. 27 (eds. Ghuysen, J.-M. & Hakenbeek, R.) 39–54 (Elsevier Science B.V., Amsterdam, 1994).
Brötz, H. et al. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30, 317–327 (1998).
Sheldrick, G.M., Jones, P.G., Kennard, O., Williams, D.H. & Smith, G.A. Structure of Vancomycin and its complex with acetyl-D-alanyl-D-alanine. Nature 271, 223–225 (1978).
McCafferty, D.G. et al. Chemistry and biology of the Ramoplanin family of peptide antibiotics. Biopolymers 66, 261–284 (2002).
Hughes, D. Exploiting genomics, genetics and chemistry to combat antibiotic resistance. Nat. Rev. Genet. 4, 432–441 (2003).
Sahl, H.-G. & Bierbaum, G. LANTIBIOTICS: Biosynthesis and biological activities of uniquely modified peptides from Gram-positive bacteria. Annu. Rev. Microbiol. 52, 41–79 (1998).
Guder, A., Wiedemann, I. & Sahl, H.-G. Posttranslationally modified bacteriocins—the lantibiotics. Biopolymers 55, 62–73 (2000).
Breukink, E. et al. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286, 2361–2364 (1999).
Breukink, E. et al. Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J. Biol. Chem. 26, 26 (2003).
Zhou, G.P. & Troy, F.A. 2nd. Characterization by NMR and molecular modeling of the binding of polyisoprenols and polyisoprenyl recognition sequence peptides: 3D structure of the complexes reveals sites of specific interactions. Glycobiology 13, 51–71 (2003).
Hsu, S.-T. et al. Mapping the targeted membrane pore formation mechanism by solution NMR: The nisin Z and lipid II interaction in SDS micelles. Biochemistry 41, 7670–7676 (2002).
Long, S.B., Casey, P.J. & Beese, L.S. Reaction path of protein farnesyltransferase at atomic resolution. Nature 419, 645–650 (2002).
Kuipers, O.P., Rollema, H.S., de Vos, W.M. & Siezen, R.J. Biosynthesis and secretion of a precursor of nisin Z by Lactococcus lactis, directed by the leader peptide of the homologous lantibiotic subtilin from Bacillus subtilis. FEBS Lett. 330, 23–27 (1993).
Wiedemann, I. et al. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276, 1772–1779 (2001).
Chan, W.C., Bycroft, B.W., Lian, L.Y. & Roberts, G.C.K. Isolation and characterisation of two degradation products derived from the peptide antibiotics nisin. FEBS Lett. 252, 29–36 (1989).
Liu, W. & Hansen, J.N. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J. Biol. Chem. 267, 25078–25085 (1992).
van Heusden, H., de Kruijff, B. & Breukink, E. Lipid II induces an overall transmembrane orientation of the pore-forming peptide lantibiotic nisin. Biochemistry 41, 12171–12178 (2002).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Johnson, B.A. & Blevins, R.A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994).
Cavanagh, J., Fairbrother, W.J., Palmer III, A.G. & Skelton, N.J. Protein NMR Spectroscopy (Academic Press, San Diego, CA, 1996).
Mishima, M. et al. Intermolecular 31P-15N and 31P-1H scalar couplings across hydrogen bonds formed between a protein and a nucleotide. J. Am. Chem. Soc. 122, 5883–5884 (2000).
Löhr, F., Mayhew, S.G. & Rüterjans, H. Detection of scalar couplings across NH...OP and OH...OP hydrogen bonds in a flavoprotein. J. Am. Chem. Soc. 122, 9289–9295 (2000).
Basus, V.J. Proton nuclear-magnetic-resonance assignments. Methods Enzymol. 177, 132–149 (1989).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Dominguez, C., Boelens, R. & Bonvin, A.M.J.J. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 (2003).
Linge, J., Williams, M.A., Spronk, C.A.E.M., Bonvin, A.M.J.J. & Nilges, M. Refinement of protein structures in explicit solvent. Proteins 50, 496–506 (2002).
Wallace, A.C., Laskowski, R.A. & Thornton, J.M. LIGPLOT: a program to generate schematic diagrams of protein ligand interactions. Protein Eng. 8, 127–134 (1995).
Smith, L. et al. Covalent structure of mutacin 1140 and a novel method for the rapid identification of lantibiotics. Eur. J. Biochem. 267, 6810–6816 (2000).
Acknowledgements
This work was in part supported through the National NMR facility at Utrecht University from the Netherlands Organization for Chemical Research (NWO-CW). A.M.J.J.B. is a recipient of a NWO Jonge Chemici grant. A.M.G.L. is supported by NWO-STW grant 349-5257. We thank E.J. Smid (NIZO-food research) for his help with the preparation of 15N-labeled nisin.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Chemical shift perturbations upon nisin–3LII complexation. (PDF 250 kb)
Rights and permissions
About this article
Cite this article
Hsu, ST., Breukink, E., Tischenko, E. et al. The nisin–lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11, 963–967 (2004). https://doi.org/10.1038/nsmb830
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb830
This article is cited by
-
Antibacterial natural products from microbial and fungal sources: a decade of advances
Molecular Diversity (2023)
-
Recombinant production of the lantibiotic nisin using Corynebacterium glutamicum in a two-step process
Microbial Cell Factories (2022)
-
New insights into the resistance mechanism for the BceAB-type transporter SaNsrFP
Scientific Reports (2022)
-
Lantibiotics: an antimicrobial asset in combating aquaculture diseases
Aquaculture International (2022)
-
Teixobactin kills bacteria by a two-pronged attack on the cell envelope
Nature (2022)