Abstract
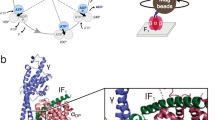
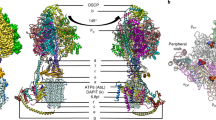
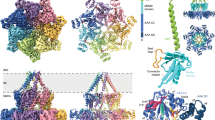
In mitochondria, the hydrolytic activity of ATP synthase is prevented by an inhibitor protein, IF1. The active bovine protein (84 amino acids) is an α-helical dimer with monomers associated via an antiparallel α-helical coiled coil composed of residues 49–81. The N-terminal inhibitory sequences in the active dimer bind to two F1-ATPases in the presence of ATP. In the crystal structure of the F1−IF1 complex at 2.8 Å resolution, residues 1–37 of IF1 bind in the αDP-βDP interface of F1-ATPase, and also contact the central γ subunit. The inhibitor opens the catalytic interface between the αDP and βDP subunits relative to previous structures. The presence of ATP in the catalytic site of the βDP subunit implies that the inhibited state represents a pre-hydrolysis step on the catalytic pathway of the enzyme.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Boyer, P.D. The ATP synthase: a splendid molecular machine. Annu. Rev. Biochem. 66, 717–749 (1997).
Walker, J.E. ATP synthesis by rotary catalysis (Nobel lecture). Angew. Chem. Int. Edn. Engl. 37, 2309–2319 (1998).
Senior, A.E., Nadanaciva, S. & Weber, J. The molecular mechanism of ATP synthesis by F1Fo-ATP synthase. Biochim. Biophys. Acta 1553, 188–211 (2002).
Collinson, I.R. et al. ATP synthase from bovine heart mitochondria. In vitro assembly of a stalk complex in the presence of F1-ATPase and in its absence. J. Mol. Biol. 242, 408–421 (1994).
Karrasch, S. & Walker, J.E. Novel features in the structure of bovine ATP synthase. J. Mol. Biol. 290, 379–384 (1999).
Rubinstein, J. & Walker, J.E. ATP synthase from Saccharomyces cerevisiae: location of the OSCP subunit in the peripheral stalk region. J. Mol. Biol. 321, 613–619 (2002).
Stock, D., Leslie, A.G.W. & Walker, J.E. Molecular architecture of the rotary motor in ATP synthase. Science 286, 1700–1705 (1999).
Gibbons, C., Montgomery, M.G., Leslie, A.G.W. & Walker, J.E. The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat. Struct. Biol. 7, 1055–1061 (2000).
Abrahams, J.P., Leslie, A.G.W., Lutter, R. & Walker, J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994).
Pullman, M.E. & Monroy, G.C. A soluble heat stable protein in mitochondria from bovine heart that inhibits ATP hydrolase activity. J. Biol. Chem. 238, 3762–3769 (1963).
Green, D.W. & Grover, G.J. The IF1 inhibitor protein of the mitochondrial F1Fo-ATPase. Biochim. Biophys. Acta 1458, 343–355 (2000).
van Raaij, M.J. et al. The ATPase inhibitor protein from bovine heart mitochondria: the minimal inhibitory sequence. Biochemistry 35, 15618–15625 (1996).
Gordon-Smith, D.J. et al. Solution structure of a C-terminal coiled-coil domain from bovine IF1: the inhibitor protein of F1-ATPase. J. Mol. Biol. 308, 325–339 (2001).
Cabezon, E., Runswick, M.J., Leslie, A.G.W. & Walker, J.E. The structure of bovine IF1, the regulatory subunit of mitochondrial F1-ATPase. EMBO J. 20, 6990–6996 (2001).
Cabezon, E., Arechaga, I., Butler, P.J.G. & Walker, J.E. Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1 . J. Biol. Chem. 275, 28353–28355 (2000).
Cabezon, E., Butler, P.J.G., Runswick, M.J. & Walker, J.E. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J. Biol. Chem. 275, 25460–25464 (2000).
Rouslin, W. Protonic inhibition of the mitochondrial oligomycin-sensitive adenosine 5′-triphosphatase in ischemic and autolyzing cardiac muscle. Possible mechanism for the mitigation of ATP hydrolysis under nonenergizing conditions. J. Biol. Chem. 258, 9657–9661 (1983).
Rouslin, W. Factors affecting the reactivation of the oligomycin-sensitive adenosine 5′-triphosphatase and the release of ATPase inhibitor protein during the re-energization of intact mitochondria from ischemic cardiac muscle. J. Biol. Chem. 262, 3472–3476 (1987).
Rouslin, W. & Broge, C.W. Regulation of mitochondrial matrix pH and adenosine 5′-triphosphatase activity during ischemia in slow heart-rate hearts. Role of Pi/H+ symport. J. Biol. Chem. 264, 15224–15229 (1989).
Mimura, H., Hashimoto, T., Yoshida, Y., Ichikawa, N. & Tagawa, K. Binding of an intrinsic ATPase inhibitor to the interface between α- and β-subunits of F1Fo-ATPase upon de-energization of mitochondria. J. Biochem. (Tokyo) 113, 350–354 (1993).
Jackson, P.J. & Harris, D.A. The mitochondrial ATP synthase inhibitor protein binds near the C terminus of the F1 β-subunit. FEBS Lett. 229, 224–228 (1988).
Power, J., Cross, R.L. & Harris, D.A. Interaction of F1-ATPase, from ox heart mitochondria with its naturally occurring inhibitor protein. Studies using radio-iodinated inhibitor protein. Biochim. Biophys. Acta 724, 128–141 (1983).
Milgrom, Y.M. When beef-heart mitochondrial F1-ATPase is inhibited by inhibitor protein a nucleotide is trapped in one of the catalytic sites. Eur. J. Biochem. 200, 789–795 (1991).
Penin, F., Di Pietro, A., Godinot, C. & Gautheron, D.C. Fate of nucleotides bound to reconstituted Fo-F1 during adenosine 5′-triphosphate synthesis activation or hydrolysis: role of protein inhibitor and hysteretic inhibition. Biochemistry 27, 8969–8974 (1988).
Pedersen, P.L., Schwerzmann, K. & Cintron, N.M. Regulation of the synthesis and hydrolysis of ATP in biological systems: role of peptide inhibitors of H+-ATPases. Curr. Top. Bioenerg. 11, 149–199 (1981).
Boyer, P.D. The binding change mechanism for ATP synthase: some probabilities and possibilities. Biochim. Biophys. Acta 1140, 215–250 (1993).
Nadanaciva, S., Weber, J., Wilke-Mounts, S. & Senior, A.E. Importance of F1-ATPase residue α-Arg376 for catalytic transition state stabilisation. Biochemistry 38, 15493–15499 (1999).
Cabezon, E., Butler, P.J., Runswick, M.J., Carbajo, R.J. & Walker, J.E. Homologous and heterologous inhibitory effects of ATPase inhibitor proteins on F1-ATPases. J. Biol. Chem. 277, 41334–41341 (2002).
Klionsky, D.J., Bruslow, W.S.A. & Simoni, R.D. In vivo evidence for the role of the ε-subunit as an inhibitor of the proton-translocating ATPase of Escherichia coli. J. Bacteriol. 160, 1055–1060 (1985).
Hausrath, A.C., Gruber, G., Matthews, B.W. & Capaldi, R.A. Structural features of the γ-subunit of the Escherichia coli F1-ATPase revealed by a 4.4 Å resolution map obtained by X-ray crystallography. Proc. Natl. Acad. Sci. USA 96, 13697–13702 (1999).
Rodgers, A.J. & Wilce, M.C. Structure of the γ-ε complex of ATP synthase. Nat. Struct. Biol. 7, 1051–1054 (2000).
Hausrath, A.C., Capaldi, R.A. & Matthews, B.W. The conformation of the ε- and γ-subunits within the Escherichia coli F1-ATPase. J. Biol. Chem. 276, 47227–47232 (2001).
Tsunoda, S.P. et al. Large conformational changes of the ε-subunit in the bacterial F1Fo ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc. Natl. Acad. Sci. USA 98, 6560–6564 (2001).
Aggeler, R., Haughton, M.A. & Capaldi, R.A. Disulfide bond formation between the COOH-terminal domain of the β-subunits and the γ- and ε-subunits of the Escherichia coli F1-ATPase. Structural implications and functional consequences. J. Biol. Chem. 270, 9185–9191 (1995).
Leslie, A.W.G. Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography Vol. 26 (Daresbury Laboratory, Warrington, UK, 1992).
Collaborative Computational Project, Number 4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Navaza, J. AmoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Braig, K., Menz, R.I., Montgomery, M.G., Leslie, A.G.W. & Walker, J.E. Structure of bovine mitochondrial F1-ATPase inhibited by Mg2+ ADP and aluminum fluoride. Structure 8, 567–573 (2000).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. A 54, 905–921 (1998).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Esnouf, R.M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. 15, 132–134 (1997).
Acknowledgements
We thank the staff of beamline ID14 at European Synchrotron Radiation Facility (ESRF), Grenoble, for help with data collection. E.C. was supported during part of this work by a European Molecular Biology Organization Fellowship and by a TMR Marie Curie Research Training Grant from the European Community.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Cabezón, E., Montgomery, M., Leslie, A. et al. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat Struct Mol Biol 10, 744–750 (2003). https://doi.org/10.1038/nsb966
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb966
This article is cited by
-
Molecular mechanism on forcible ejection of ATPase inhibitory factor 1 from mitochondrial ATP synthase
Nature Communications (2023)
-
Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi
Nature Communications (2020)
-
Catalytic robustness and torque generation of the F1-ATPase
Biophysical Reviews (2017)
-
Diabetes-induced abnormalities of mitochondrial function in rat brain cortex: the effect of n-3 fatty acid diet
Molecular and Cellular Biochemistry (2017)
-
The shrimp mitochondrial FoF1-ATPase inhibitory factor 1 (IF1)
Journal of Bioenergetics and Biomembranes (2015)