Abstract
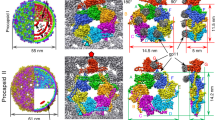
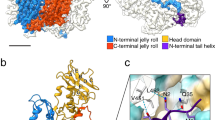
Three-dimensional structures of the double-stranded DNA bacteriophage φ29 scaffolding protein (gp7) before and after prohead assembly have been determined at resolutions of 2.2 and 2.8 Å, respectively. Both structures are dimers that resemble arrows, with a four-helix bundle composing the arrowhead and a coiled coil forming the tail. The structural resemblance of gp7 to the yeast transcription factor GCN4 suggests a DNA-binding function that was confirmed by native gel electrophoresis. DNA binding to gp7 may have a role in mediating the structural transition from prohead to mature virus and scaffold release. A cryo-EM analysis indicates that gp7 is arranged inside the capsid as a series of concentric shells. The position of the higher density features in these shells correlates with the positions of hexamers in the equatorial region of the capsid, suggesting that gp7 may regulate formation of the prolate head through interactions with these hexamers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Anderson, D. & Reilly, B. Morphogenesis of bacteriophage φ29. In Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics (ed. Sonenshein, A.L., Hoch, J.A. & Losick, R.) 859–867 (American Society for Microbiology, Washington, DC, 1993).
Tao, Y. et al. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 95, 431–437 (1998).
Hagen, E.W., Reilly, B.E., Tosi, M.E. & Anderson, D.L. Analysis of gene function of bacteriophage φ29 of Bacillus subtilis: identification of cistrons essential for viral assembly. J. Virol. 19, 501–517 (1976).
Guo, P. et al. Regulation of the phage φ29 prohead shape and size by the portal vertex. Virology 183, 366–373 (1991).
Guo, P., Peterson, C. & Anderson, D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J. Mol. Biol. 197, 229–236 (1987).
Bjornsti, M.A., Reilly, B.E. & Anderson, D.L. Morphogenesis of bacteriophage φ29 of Bacillus subtilis: oriented and quantized in vitro packaging of DNA-protein gp3. J. Virol. 45, 383–396 (1983).
Crick, F.H.C. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 6, 689–697 (1953).
Branden, C. & Tooze, J. Introduction to Protein Structure (Garland Publishing, New York and London, 1991).
Lee, C.S. & Guo, P. Sequential interactions of structural proteins in phage φ29 procapsid assembly. J. Virol. 69, 5024–5032 (1995).
Fane, B.A. & Prevelige, P.E. Jr. Mechanism of scaffolding-assisted viral assembly. Adv. Prot. Chem. (in the press).
Sun, Y. et al. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol. 297, 1195–1202 (2000).
Thuman-Commike, P.A. et al. Identification of additional coat-scaffolding interactions in a bacteriophage P22 mutant defective in maturation. J. Virol. 74, 3871–3873 (2000).
Jiang, W. et al. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 10, 131–135 (2003).
Peterson, C. et al. Composition and mass of the bacteriophage φ29 prohead and virion. J. Struct. Biol. 135, 18–25 (2001).
Parker, M.H., Brouillette, C.G. & Prevelige, P.E. Jr. Kinetic and calorimetric evidence for two distinct scaffolding protein binding populations within the bacteriophage P22 procapsid. Biochemistry 40, 8962–8970 (2001).
Aebi, U. et al. The transformation of τ particles into T4 heads. II. Transformations of the surface lattice and related observations on form determination. J. Supramol. Struct. 2, 253–275 (1974).
Ellenberger, T.E., Brandl, C.J., Struhl, K. & Harrison, S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α-helices: crystal structure of the protein–DNA complex. Cell 71, 1223–1237 (1992).
Wichitwechkarn, J., Bailey, S., Bodley, J.W. & Anderson, D. Prohead RNA of bacteriophage φ29: size, stoichiometry and biological activity. Nucleic Acids Res. 17, 3459–3468 (1989).
Grimes, S. & Anderson, D. The bacteriophage φ29 packaging proteins supercoil the DNA ends. J. Mol. Biol. 266, 901–914 (1997).
Cowtan, K.D. & Main, P. Improvement of macromolecular electron-density maps by the simultaneous application of real and reciprocal space constraints. Acta Crystallogr. D 49, 148–157 (1993).
Cowtan, K.D. 'dm': an automated procedure for phase improvement by density modification. in Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography Vol. 31 (Daresbury Laboratory, Warrington, UK, 1994), 34–38.
Rossmann, M.G. The molecular replacement method. Acta Crystallogr. A 46, 73–82 (1990).
Navaza, J. AMoRe—an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994).
Brünger, A.T. et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Acknowledgements
We thank P. Jardine for helpful discussions and encouragement, and S. Wilder and C. Towell for help in preparation of the manuscript. The work was supported by a US National Science Foundation grant to M.G.R., National Institutes of Health grants to D.L.A. and a National Institutes of Health postdoctoral fellowship to M.C.M. We are also grateful for a grant from the Keck Foundation that provided for the purchase of a Philips CM300 FEG electron microscope, as well as for a Purdue University reinvestment grant to support structural biology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Morais, M., Kanamaru, S., Badasso, M. et al. Bacteriophage φ29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Mol Biol 10, 572–576 (2003). https://doi.org/10.1038/nsb939
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb939