Abstract
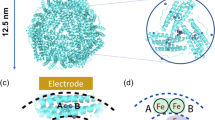
The first crystal structure of a native di-iron center in an iron-storage protein (bacterio)ferritin is reported. The protein, isolated from the anaerobic bacterium Desulfovibrio desulfuricans, has the unique property of having Fe-coproporphyrin III as its heme cofactor. The three-dimensional structure of this bacterioferritin was determined in three distinct catalytic/redox states by X-ray crystallography (at 1.95, 2.05 and 2.35 Å resolution), corresponding to different intermediates of the di-iron ferroxidase site. Conformational changes associated with these intermediates support the idea of a route for iron entry into the protein shell through a pore that passes through the di-iron center. Molecular surface and electrostatic potential calculations also suggest the presence of another ion channel, distant from the channels at the three- and four-fold axes proposed as points of entry for the iron atoms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Crichton, R. Intracellular iron storage and biomineralization. in Inorganic Biochemistry of Iron Metabolism: From Molecular Mechanisms to Clinical Consequences 2nd edn. (eds. Boelaert, J.R. et al.) 133–165 (John Wiley, Chichester; 2001).
Harrison, P.M., Hempstead, P.D., Artymiuk, P.J. & Andrews, S.C. Structure-function relationships in the ferritins. in Metal Ions in Biological Systems Vol. 35 (eds. Siegel, A. & Siegel, H.) 435–477 (Marcel Dekker, New York; 1998).
Banyard, S.H., Stammers, D.K. & Harrison, P.M. Electron density map of apoferritin at 2.8-Å resolution. Nature 271, 282–284 (1978).
Romão, C.V. et al. A bacterioferritin from the strict anaerobe Desulfovibrio desulfuricans ATCC 27774. Biochemistry 39, 6841–6849 (2000).
Romão, C.V. et al. Iron-coproporphyrin III is a natural cofactor in bacterioferritin from the anaerobic bacterium Desulfovibrio desulfuricans. FEBS Lett. 480, 213–216 (2000).
da Costa, P.N. et al. The genetic organization of Desulfovibrio desulphuricans ATCC 27774 bacterioferritin and rubredoxin-2 genes: involvement of rubredoxin in iron metabolism. Mol. Microbiol. 41, 217–227 (2001).
Trikha, J., Theil, E.C. & Allewell, N.M. High resolution crystal structures of amphibian red-cell L ferritin: potential roles for structural plasticity and solvation in function. J. Mol. Biol. 248, 949–967 (1995).
Gallois, B. et al. X-ray structure of recombinant horse L chain apoferritin at 2.0 Å resolution: implications for stability and function. J. Biol. Inorg. Chem. 2, 360–367 (1997).
Hempstead, P.D. et al. Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution. J. Mol. Biol. 268, 424–448 (1997).
Granier, T. et al. Structure of mouse L-chain ferritin at 1.6 Å resolution. Acta Crystallogr. D 57, 1491–1497 (2001).
Stillman, T.J. et al. The high-resolution X-ray crystallographic structure of the ferritin (EcFtnA) of Escherichia coli; comparison with human H ferritin (HuHF) and the structures of the Fe3+ and Zn2+ derivatives. J. Mol. Biol. 307, 587–603 (2001).
Frolow, F. & Kalb (Gilboa), A.J. Cytochrome b1 – bacterioferritin. in Handbook of Metalloproteins Vol. 2 (eds. Messerschmidt, A., Huber, R., Poulos, T. & Wieghardt, K.) 782–790 (Wiley, UK; 2001).
Ilari, A., Stefanini, S., Chiancone, E. & Tsernoglou, D. The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nat. Struct. Biol. 7, 38–43 (2000).
Frolow, F., Kalb (Gilboa), A.J. & Yariv, J. Structure of a unique two-fold symmetric heme-binding site. Nat. Struct. Biol. 1, 453–460 (1994).
Dautant, A. et al. Structure of a monoclinic crystal from of cyctochrome b1 (bacterioferritin) from E. coli. Acta Crystallogr. D 54, 16–24 (1998).
Cobessi, D. et al. The 2.6 Å resolution structure of Rhodobacter capsulatus bacterioferritin with metal-free dinuclear site and heme iron in a crystallographic 'special position'. Acta Crystallogr. D 58, 29–38 (2002).
Coelho, A.V. et al. Structure determination of bacterioferritin from Desulfovibrio desulfuricans by the MAD method at the Fe K-edge. Acta Crystallogr. D 57, 326–329 (2001).
Theil, E.C. Ferritin. in Handbook of Metalloproteins Vol. 2 (eds. Messerschmidt, A., Huber, R., Poulos, T. & Wieghardt, K.) 771–781 (Wiley, UK; 2001).
Rosenzweig, A.C. et al. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins 29, 141–152 (1997).
Tong, W. et al. Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography. Biochemistry 37, 5840–5848 (1998).
Gomes, C.M., Gall, J.L., Xavier, A.V. & Teixeira, M. Could a diiron-containing four-helix-bundle protein have been a primitive oxygen reductase? Chembiochem 2, 583–587 (2001).
Kurtz, D.M. Jr. Structural similarity and functional diversity in diiron-oxo proteins. J. Biol. Inorg. Chem. 2, 159–167 (1997).
Yang, X. et al. The iron oxidation and hydrolysis chemistry of Escherichia coli bacterioferritin. Biochemistry 39, 4915–4923 (2000).
Bou-Abdallah, F. et al. Iron detoxification properties of Escherichia coli bacterioferritin. Attenuation of oxyradical chemistry. J. Biol. Chem. 277, 37064–37069 (2002).
Douglas, T. & Ripoll, D.R. Calculated electrostatic gradients in recombinant human H-chain ferritin. Protein Sci. 7, 1083–1091 (1998).
Chasteen, N.D. & Harrison, P.M. Mineralization in ferritin: an efficient means of iron storage. J. Struct. Biol. 126, 182–194 (1999).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Cowtan, K.D. & Main, P. Miscellaneous algorithms for density modification. Acta Crystallogr. D 54, 487–493 (1998).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Kleywegt, G.J., Zou, J.Y., Kjeldgaard, M. & Jones, T.A. Model building and computer graphics. in International Tables for Crystallography, Vol. F, Crystallography of biological macromolecules (eds. Rossmann, M.G. & Arnold, E.) 353–356 (Kluwer Academic Publishers, The Netherlands; 2001).
Read, R.J. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42, 140–149 (1986).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Murshodov, G.N., Vagin, A.A., Lebedev, A., Wilson, K.S. & Dodson, E.J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D 55, 247–255 (1999).
Sheldrick, G.M. & Schneider, T.R. SHELXL: high-resolution refinement. Methods Enzymol. 277, 319–343 (1997).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK — a program to check the stereochemical quality of proteins. J. Appl. Crystallogr. 26, 283–291 (1993).
Kraulis, P.J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E.A. & Murphy, M.E.P. Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994).
Esnouf, R.M. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55, 938–940 (1999).
Sanner, M.F., Spehner, J.-C. & Olson, A.J. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305–320 (1996).
Bashford, D. An object-oriented programming suite for electrostatic effects in biological molecules. Scientific computing in object-oriented parallel environments. in Lecture Notes in Computer Science Vol. 1343 (eds. Ishikawa, Y., Oldehoeft, R.R., Reynders, J.V.W. & Tholburn, M.) 233–240 (Springer, Berlin; 1997).
Acknowledgements
This work was supported by PRAXIS XXI and POCTI grants. We thank the ESRF, Grenoble (France) for the provision of synchrotron radiation and for a grant to S.M.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Macedo, S., Romão, C., Mitchell, E. et al. The nature of the di-iron site in the bacterioferritin from Desulfovibrio desulfuricans. Nat Struct Mol Biol 10, 285–290 (2003). https://doi.org/10.1038/nsb909
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb909
This article is cited by
-
Genomic insight into iron acquisition by sulfate-reducing bacteria in microaerophilic environments
BioMetals (2023)
-
New Insights into the Role of Ferritin in Iron Homeostasis and Neurodegenerative Diseases
Molecular Neurobiology (2021)
-
Metal homeostasis and resistance in bacteria
Nature Reviews Microbiology (2017)
-
Ferritins: furnishing proteins with iron
JBIC Journal of Biological Inorganic Chemistry (2016)
-
Structure/function correlations over binuclear non-heme iron active sites
JBIC Journal of Biological Inorganic Chemistry (2016)