Abstract
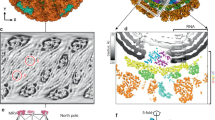
Reovirus is an icosahedral, double-stranded (ds) RNA virus that uses viral polymerases packaged within the viral core to transcribe its ten distinct plus-strand RNAs. To localize these polymerases, the structure of the reovirion was refined to a resolution of 7.6 Å by cryo-electron microscopy (cryo-EM) and three-dimensional (3D) image reconstruction. X-ray crystal models of reovirus proteins, including polymerase λ3, were then fitted into the density map. Each copy of λ3 was found anchored to the inner surface of the icosahedral core shell, making major contacts with three molecules of shell protein λ1 and overlapping, but not centering on, a five-fold axis. The overlap explains why only one copy of λ3 is bound per vertex. λ3 is furthermore oriented with its transcript exit channel facing a small channel through the λ1 shell, suggesting how the nascent RNA is passed into the large external cavity of the pentameric capping enzyme complex formed by protein λ2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nibert, M.L., Schiff, L.A. & Fields, B.N. Reoviruses and their replication. in Fields Virology (eds. Fields, B.N., Knipe, D.M. & Howley, P.M.) 1679–1728 (Raven, Philadelphia, 2001).
Dryden, K.A. et al. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 122, 1023–1041 (1993).
Liemann, S., Chandran, K., Baker, T.S., Nibert, M.L. & Harrison, S.C. Structure of the reovirus membrane-penetration protein, μ1, in a complex with its protector protein, σ3. Cell 108, 283–295 (2002).
Furlong, D.B., Nibert, M.L. & Fields, B.N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62, 246–256 (1988).
Strong, J.E., Leone, G., Duncan, R., Sharma, R.K. & Lee, P.W.K. Biochemical and biophysical characterization of the reovirus cell attachment protein σ1: evidence that it is a homotrimer. Virology 184, 23–32 (1991).
Larson, S.M., Antczak, J.B. & Joklik, W.K. Reovirus exists in the form of 13 particle species that differ in their content of protein σ1. Virology 201, 303–311 (1994).
Chappell, J.D., Porta, A.E., Dermody, T.S. & Stehle, T. Crystal structure of reovirus attachment protein σ1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21, 1–11 (2002).
Sturzenbecker, L.J., Nibert, M., Furlong, D. & Fields, B.N. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61, 2351–2361 (1987).
Chandran, K., Farsetta, D.L. & Nibert, M.L. Strategy for nonenveloped virus entry: a hydrophobic conformer of reovirus penetration protein μ1 mediates membrane disruption. J. Virol. 76, 9920–9933 (2002).
Chandran, K., Parker, J.S.L., Ehrlich, M., Kirchhausen, Y. & Nibert, M.L. The δ region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J. Virol. (in the press).
Reinisch, K.M., Nibert, M.L. & Harrison, S.C. Structure of the reovirus core at 3.6 Å resolution. Nature 404, 960–967 (2000).
Samuel, C.E. Reoviruses and the interferon system. Curr. Top. Microbiol. Immunol. 233, 125–145 (1998).
Cullen, B.R. RNA interference: antiviral defense and genetic tool. Nat. Immunol. 3, 597–599 (2002).
Coombs, K.M. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology 243, 218–228 (1998).
Dryden, K.A. et al. Internal structures containing transcriptase-related proteins in top component particles of mammalian orthoreovirus. Virology 245, 33–46 (1998).
Drayna, D. & Fields, B.N. Activation and characterization of the reovirus transcriptase: genetic analysis. J. Virol. 41, 110–118 (1982).
Starnes, M.C. & Joklik, W.K. Reovirus protein λ3 is a poly(C)-dependent poly(G) polymerase. Virology 193, 356–366 (1993).
Tao, Y., Farsetta, D.L., Nibert, M.L. & Harrison, S.C. RNA synthesis in a cage—structural studies of reovirus polymerase λ3. Cell 111, 733–745 (2002).
Yin, P., Cheang, M. & Coombs, K.M. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J. Virol. 70, 1223–1227 (1996).
Noble, S. & Nibert, M.L. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 71, 7728–7735 (1997).
Noble, S. & Nibert, M.L. Characterization of an ATPase activity in reovirus cores and its genetic association with core-shell protein λ1. J. Virol. 71, 2182–2191 (1997).
Bisaillon, M., Bergeron, J. & Lemay, G. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus λ1 protein. J. Biol. Chem. 272, 18298–18303 (1997).
Bisaillon, M. & Lemay, G. Characterization of the reovirus λ1 protein RNA 5′-triphosphatase activity. J. Biol. Chem. 272, 29954–29957 (1997).
Furuichi, Y., Muthukrishnan, S., Tomasz, J. & Shatkin, A.J. Caps in eukaryotic mRNAs: mechanism of formation of reovirus mRNA 5′-terminal m7GpppGm-C. Prog. Nucleic Acid Res. Mol. Biol. 19, 3–20 (1976).
Cleveland, D.R., Zarbl, H. & Millward, S. Reovirus guanylyltransferase is L2 gene product λ2. J. Virol. 60, 307–311 (1986).
Mao, Z.X. & Joklik, W.K. Isolation and enzymatic characterization of protein λ2, the reovirus guanylyltransferase. Virology 185, 377–386 (1991).
Fausnaugh, J. & Shatkin, A.J. Active site localization in a viral mRNA capping enzyme. J. Biol. Chem. 265, 7669–7672 (1990).
Luongo, C.L., Contreras, C.M., Farsetta, D.L. & Nibert, M.L. Binding site for S-adenosyl-L-methionine in a central region of mammalian reovirus λ2 protein. Evidence for activities in mRNA cap methylation. J. Biol. Chem. 273, 23773–23780 (1998).
Luongo, C.L., Reinisch, K.M., Harrison, S.C. & Nibert, M.L. Identification of the guanylyltransferase region and active site in reovirus mRNA capping protein λ2. J. Biol. Chem. 275, 2804–2810 (2000).
Gillies, S., Bullivant, S. & Bellamy, A.R. Viral RNA polymerases: electron microscopy of reovirus reaction cores. Science 174, 694–696 (1971).
Bartlett, N.M., Gillies, S.C., Bullivant, S. & Bellamy, A.R. Electron microscopy study of reovirus reaction cores. J. Virol. 14, 315–326 (1974).
Yeager, M., Weiner, S. & Coombs, K.M. Transcriptionally active reovirus core particles visualized by electron cryo-microscopy and image reconstruction. Biophys. J. 70, A116 (1996).
Furuichi, Y. & Shatkin, A.J. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55, 135–184 (2000).
Shuman, S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66, 1–40 (2001).
Prasad, B.V.V. et al. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382, 471–473 (1996).
Pesavento, J.B., Lawton, J.A., Estes, M.K. & Prasad, B.V.V. The reversible condensation and expansion of the rotavirus genome. Proc. Natl. Acad. Sci. USA 98, 1381–1386 (2001).
Gouet, P. et al. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97, 481–490 (1999).
Grimes, J.M. et al. The atomic structure of the bluetongue virus core. Nature 395, 470–478 (1998).
Zhang, H. et al. Visualization of protein-RNA interactions in cytoplasmic polyhedrosis virus. J. Virol. 73, 1624–1629 (1999).
Baker, T.S., Olson, N.H. & Fuller, S.D. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63, 862–922 (1999).
Rossmann, M.G., Bernal, R. & Pletnev, S.V. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136, 190–200 (2001).
Chacón, P. & Wriggers, W. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 317, 375–384 (2002).
Farsetta, D.L., Chandran, K. & Nibert, M.L. Transcriptional activities of reovirus RNA polymerase in recoated cores. Initiation and elongation are regulated by separate mechanisms. J. Biol. Chem. 275, 39693–39701 (2000).
Joklik, W.K. The reovirus particle. In The Reoviridae (ed. Joklik, W.K.) 9–78 (Plenum, New York, 1983).
Shatkin, A.J. & Kozak, M. Biochemical aspects of reovirus transcription and translation. In The Reoviridae (ed. Joklik, W.K.) 79–106 (Plenum, New York, 1983).
Cheetham, G.M. & Steitz, T.A. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10, 117–123 (2000).
Spencer, S.M., Sgro, J.-Y., Dryden, K.A., Baker, T.S. & Nibert, M.L. IRIS explorer software for radial-depth cueing reovirus particles and other macromolecular structures determined by cryoelectron microscopy and image reconstruction. J. Struct. Biol. 120, 11–21 (1997).
Luongo, C.L. et al. Loss of activities for mRNA synthesis accompanies loss of λ2 spikes from reovirus cores: an effect of λ2 on λ1 shell structure. Virology 296, 24–38 (2002).
Diprose, J.M. et al. Translocation portals for the substrates and products of a viral transcription complex: the bluetongue virus core. EMBO J. 20, 7229–7239 (2001).
Lawton, J.A., Estes, M.K. & Prasad, B.V.V. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4, 118–121 (1997).
Le Blois, H., French, T., Mertens, P.P., Burroughs, J.N. & Roy, P. The expressed VP4 protein of bluetongue virus binds GTP and is the candidate guanylyl transferase of the virus. Virology 189, 757–761 (1992).
Baker, T.S. & Cheng, R.H. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116, 120–130 (1996).
Bowman, V.D. et al. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76, 12250–12258 (2002).
van Heel, M. et al. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Rev. Biophys. 33, 371–424 (2000).
Esnouf, R.M. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15, 132–134 (1997).
Merritt, E.A. & Bacon, D.J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Grimes, J.M. et al. An atomic model of the outer layer of the bluetongue virus core derived from X-ray crystallography and electron cryomicroscopy. Structure 5, 885–893 (1997).
Acknowledgements
We are especially grateful to S. Harrison and Y. Tao for providing the coordinates of the λ3 crystal structure before publication and also for discussions and comments on the manuscript. We also thank W. Zhang, C. Xiao, R. Ashmore, J. Johnson, A. McGough, R. Bernal, M. Sherman, M. Rossmann, P. Chacón and B. Bahlke for helpful discussions; V. Bowman, A. Simpson, P. Leiman, Y. Tao and J. Zhiu for assistance with figures; K. Reinisch and S. Liemann for providing crystal coordinates and discussion; and L. Szpankowski for digitizing micrographs. Work was supported in part by grants from the US National Institutes of Health to T.S.B. and M.L.N., a shared equipment grant from the US National Science Foundation to T.S.B., a Keck Foundation award to the Purdue Structural Biology group for purchase of the CM300 FEG microscope and a Purdue University reinvestment grant to the Structural Biology group. S.B.W. was additionally supported by the Purdue Biophysics Training Grant and a Purdue Research Foundation Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, X., Walker, S., Chipman, P. et al. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6 Å. Nat Struct Mol Biol 10, 1011–1018 (2003). https://doi.org/10.1038/nsb1009
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb1009
This article is cited by
-
Assembly intermediates of orthoreovirus captured in the cell
Nature Communications (2020)
-
Detection of mammalian orthoreovirus type-3 (Reo-3) infections in mice based on serotype-specific hemagglutination protein sigma-1
Virology Journal (2018)
-
In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus
Nature (2015)
-
The VP2 protein of grass carp reovirus (GCRV) expressed in a baculovirus exhibits RNA polymerase activity
Virologica Sinica (2014)
-
Structural insights into the coupling of virion assembly and rotavirus replication
Nature Reviews Microbiology (2012)