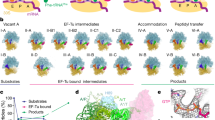
Abstract
Aminoacyl-tRNAs (aa-tRNAs) are delivered to the ribosome as part of the ternary complex of aa-tRNA, elongation factor Tu (EF-Tu) and GTP. Here, we present a cryo-electron microscopy (cryo-EM) study, at a resolution of ∼9 Å, showing that during the incorporation of the aa-tRNA into the 70S ribosome of Escherichia coli, the flexibility of aa-tRNA allows the initial codon recognition and its accommodation into the ribosomal A site. In addition, a conformational change observed in the GTPase-associated center (GAC) of the ribosomal 50S subunit may provide the mechanism by which the ribosome promotes a relative movement of the aa-tRNA with respect to EF-Tu. This relative rearrangement seems to facilitate codon recognition by the incoming aa-tRNA, and to provide the codon-anticodon recognition-dependent signal for the GTPase activity of EF-Tu. From these new findings we propose a mechanism that can explain the sequence of events during the decoding of mRNA on the ribosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rodnina, M.V., Pape, T., Fricke, R., Kuhn, L. & Wintermeyer, W. Initial binding of the elongation factor Tu·GTP·aminoacyl-tRNA complex preceding codon recognition on the ribosome. J. Biol. Chem. 271, 646–652 (1996).
Berchtold, H. et al. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature 365, 126–132 (1993).
Kjeldgaard, M., Nissen, P., Thirup, S. & Nyborg, J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1, 35–50 (1993).
Dell, V.A., Miller, D.L. & Johnson, A.E. Effects of nucleotide- and aurodox-induced changes in elongation factor Tu conformation upon its interaction with aminoacyl-tRNA. A fluorescence study. Biochemistry 29, 1757–1763 (1990).
Kaziro, Y. The role of guanosine-5′-triphosphate in polypeptide chain elongation. Biochim. Biophys. Acta 505, 95–127 (1978).
Stark, H. et al. Visualization of elongation factor Tu on Escherichia coli ribosome. Nature 389, 403–406 (1997).
Agrawal, R.K. et al. Visualization of tRNA movements on the Escherichia coli ribosome during the elongation cycle. J. Cell Biol. 150, 447–459 (2000).
Valle, M. et al. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 21, 3557–3567 (2002).
Stark, H. et al. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 9, 849–854 (2002).
Mohr, D., Wintermeyer, W. & Rodnina, M.V. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry 41, 12520–12528 (2002).
Rodnina, M.V. & Wintermeyer, W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanism. Annu. Rev. Biochem. 70, 415–435 (2001).
Thompson, R.C. & Stone, P.J. Proofreading of the codon-anticodon interaction on ribosomes. Proc. Natl. Acad. Sci. USA 74, 198–202 (1977).
Ruusala, T., Ehrenberg, M. & Kurland, C.G. Is there proofreading during polypeptide synthesis? EMBO J. 1, 415–435 (1982).
Pape, T., Wintermeyer, W. & Rodnina, M.V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of E. coli ribosome. EMBO J. 17, 7490–7497 (1998).
Rodnina, M.V., Fricke, R. & Wintermeyer, W. Transient conformational states of aminoacyl-tRNA during ribosome binding catalyzed by elongation factor Tu. Biochemistry 33, 12267–12275 (1994).
Rodnina, M.V., Fricke, R., Kohn, L. & Wintermeyer, W. Codon-dependent conformational change of elongation factor Tu preceding GTP hydrolysis on the ribosome. EMBO J. 14, 2613–2619 (1995).
Piepenburg, O. et al. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry 39, 1734–1738 (2000).
Wolf, H., Chinali, G. & Parmeggiani, A. Mechanism of the inhibition of protein synthesis by kirromycin. Eur. J. Biochem. 75, 67–75 (1977).
Parmeggiani, A. & Stewart, G.W. Mechanism of action of kirromycin-like antibiotics. Annu. Rev. Microbiol. 39, 557–577 (1985).
Wimberly, B. et al. Structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000).
Ogle, J.M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001).
Abel, K.M., Yoder, M.D., Hilgenfeld, R. & Jurnak, F. An α to β conformational switch in EF-Tu. Structure 4, 1153–1159 (1996).
Polekhina, G. et al. Helix unwinding in the effector region of elongation factor EF-Tu-GDP. Structure 4, 1141–1151 (1996).
Hilgenfeld, R., Mesters, J. & Hogg, T. Insights into the GTPase mechanism of EF-Tu from structural studies. In The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions (eds. Garret, R.A. et al.) 347–357 (ASM Press, Washington, DC, 2000).
Vogeley, L., Palm, G.J., Mesters, J.R. & Hilgenfeld, R. Conformational change of elongation factor Tu (EF-Tu) induced by antibiotic binding. J. Biol. Chem. 276, 17149–17155 (2001).
Ban, N., Nissen, P., Hansen, J., Moore, P.B. & Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920 (2000).
Moazed, D., Robertson, J.M. & Noller, H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S rRNA. Nature 334, 362–364 (1988).
Hausner, T.P., Atmadja, J. & Nierhaus, K.H. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie 69, 911–923 (1987).
Swart, G.W.M. & Parmeggiani, A. Effects of the mutation glycine-222→aspartic acid on the functions of elongation factor Tu. Biochemistry 26, 2047–2054 (1987).
Vorstenbosch, E., Pape, T., Rodnina, M.V., Kraal, B. & Wintermeyer, W. The G222 mutation in elongation factor Tu inhibits the codon-induced conformational changes leading to GTPase activation on the ribosome. EMBO J. 15, 6766–6774 (1996).
Robertus, J.D. et al. Structure of yeast phenylananine tRNA at 3 Å resolution. Nature 250, 546–551 (1974).
Sussman, J.L., Holbrook, S.R., Warrant, R.W., Church, G.M. & Kim, S.H. Crystal structure of yeast phenylalanine transfer RNA. I. Crystallographic refinement. Science 123, 607–630 (1978).
Harvey, S.C., Prabhakaran, M., Mao, B. & McCammon, J.A. Phenylalanine transfer RNA: molecular dynamics simulations. Science 223, 1189–1191 (1984).
O'Connor, M. & Dahlberg, A.E. The involvement of two distinct regions of 23S ribosomal RNA in tRNA selection. J. Mol. Biol. 254, 838–847 (1995).
Hirsh, D. Tryptophan transfer RNA as the UGA suppressor. J. Mol. Biol. 58, 439–458 (1971).
Yarus, M. & Smith, D. tRNA on the ribosome: a Waggle theory. In tRNA: Structure, Biosynthesis, and Function (eds. Söll, D. & RajBahandary, U.) 443–468 (American Society for Microbiology, Washington, DC, 1995).
Hausner, T.P., Atmadja, J. & Nierhaus, K.H. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie 69, 911–923 (1987).
Wriggers, W., Agrawal, R.K., Drew, D.L., McCammon, A. & Frank, J. Domains motions of EF-G bound to the ribosome: insights from a hand-shaking between multi-resolution structures. Biophys. J. 79, 1670–1678 (2000).
Wimberly, B.T., Guymon, R., McCutcheon, J.P., White, S.W. & Ramakrishnan, V. A detailed view of a ribosomal active site: the structure of the L11-RNA complex. Cell 97, 491–502 (1999).
Vazquez, D. Inhibitors of Protein Synthesis (Springer, New York, 1979).
Saarma, U., Remme, J., Ehrenberg, M. & Bilgin, N. An A to U transversion at position 1067 of 23S rRNA from Escherichia coli impairs EF-Tu and EF-G function. J. Mol. Biol. 272, 327–335 (1997).
O'Connor, M. & Dahlberg, A.E. The involvement of two distinct regions of 23S ribosomal RNA in tRNA selection. J. Mol. Biol. 254, 838–847 (1995).
Zavialov, A.V., Buckingham, R.H. & Ehrenberg, M. A posttermination ribosomal complex is the guanine exchange factor for peptide release factor RF3. Cell 107, 1–20. (2001).
Rosenthal, P. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single particle electron cryomicroscopy. J. Mol. Biol., in press.
Jones, T.A., Zhou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47, 110–119 (1991).
Cornell, W.D. et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995).
Acknowledgements
We thank M. Yarus for a discussion and helpful suggestions and M. Watters for assistance with the illustrations. This work was supported by the Howard Hughes Medical Institute (J.F.), by grants from the US National Institutes of Health (J.F. and S.C.H.) and the US National Science Foundation (J.F.), by the Swedish Foundation for Strategic Research and the Swedish Research Council (A.V.Z. and M.E.), by an Ole Romer research grant from the Danish Research Council and the EMBO Young Investigator Program (P.N.) and by a scholarship from Novo Nordisk/Novozymes (R.N.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Video 1.
Accommodation process on the 70S ribosome.By alternating between cryoEM maps of (i)the 70S ribosome with the stalled ternary complex (70S ·fMet-tRNA fMet ·Phe-tRNA Phe ·EF-Tu ·GDP ·kir)and (ii)the complex bearing the accommodated A-site tRNA (70S ·tRNA fMet ·MP-tRNA Phe),the dynamic transition during the incorporation of the aa-tRNA can be observed.In the rendering,the segmented and differently colored volumes allow the ribosomal subunits (large subunit in blue,small subunit in yellow)and the tRNAs in P (green)and E sites (orange)to be distinguished. The A/T-and A-site tRNAs are depicted in the same color (magenta),so that the trajectory of the tRNA can be more easily followed during the accommodation. (MOV 2802 kb)
Supplementary Video 2.
tRNA movement and conformational change during accommodation. (MOV 3670 kb)
Rights and permissions
About this article
Cite this article
Valle, M., Zavialov, A., Li, W. et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Mol Biol 10, 899–906 (2003). https://doi.org/10.1038/nsb1003
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb1003
This article is cited by
-
Geometric alignment of aminoacyl-tRNA relative to catalytic centers of the ribosome underpins accurate mRNA decoding
Nature Communications (2023)
-
tRNA tracking for direct measurements of protein synthesis kinetics in live cells
Nature Chemical Biology (2018)
-
Recurring RNA structural motifs underlie the mechanics of L1 stalk movement
Nature Communications (2017)
-
The ribosome moves: RNA mechanics and translocation
Nature Structural & Molecular Biology (2017)
-
How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome
Nature Communications (2016)