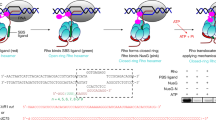
Abstract
In bacteriophage l, transcription elongation is regulated by the N protein, which binds a nascent mRNA hairpin (termed boxB) and enables RNA polymerase to read through distal terminators. We have examined the structure, energetics and in vivo function of a number of N–boxB complexes derived from in vitro protein selection. Trp18 fully stacks on the RNA loop in the wild-type structure, and can become partially or completely unstacked when the sequence context is changed three or four residues away, resulting in a recognition interface in which the best binding residues depend on the sequence context. Notably, in vivo antitermination activity correlates with the presence of a stacked aromatic residue at position 18, but not with N–boxB binding affinity. Our work demonstrates that RNA polymerase responds to subtle conformational changes in cis-acting regulatory complexes and that approximation of components is not sufficient to generate a fully functional transcription switch.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
von Hippel, P.H. An integrated model of the transcription complex in elongation, termination, and editing. Science 281, 660–665 (1998).
Yarnell, W.S. & Roberts, J.W. Mechanism of intrinsic transcription termination and antitermination. Science 284, 611–615 (1999).
Uptain, S.M., Kane, C.M. & Chamberlin, M.J. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66, 117–172 (1997).
Zorio, D.A. & Bentley, D.L. Transcription elongation: the 'Foggy' is lifting... Curr. Biol. 11, R144–R146 (2001).
Maniatis, T. & Reed, R. An extensive network of coupling among gene expression machines. Nature 416, 499–506 (2002).
Roberts, J.W. Termination factor for RNA synthesis. Nature 224, 1168–1174 (1969).
Greenblatt, J. Positive control of endolysin synthesis in vitro by the gene N protein of phage λ. Proc. Natl. Acad. Sci. USA 69, 3606–3610 (1972).
Salstrom, J.S. & Szybalski, W. Coliphage λnutL−: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J. Mol. Biol. 124, 195–221 (1978).
Greenblatt, J., Nodwell, J.R. & Mason, S.W. Transcriptional antitermination. Nature 364, 401–406 (1993).
Friedman, D.I. & Court, D.L. Transcription antitermination: the lambda paradigm updated. Mol. Microbiol. 18, 191–200 (1995).
Weisberg, R.A. & Gottesman, M.E. Processive antitermination. J. Bacteriol. 181, 359–367 (1999).
Van Gilst, M.R. & von Hippel, P.H. Assembly of the N-dependent antitermination complex of phage lambda: NusA and RNA bind independently to different unfolded domains of the N protein. J. Mol. Biol. 274, 160–173 (1997).
Mogridge, J., Mah, T.F. & Greenblatt, J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 9, 2831–2845 (1995).
Zhou, Y. et al. Interactions of an Arg-rich region of transcription elongation protein NusA with NUT RNA: implications for the order of assembly of the lambda N antitermination complex in vivo. J. Mol. Biol. 310, 33–49 (2001).
Murakami, K.S., Masuda, S., Campbell, E.A., Muzzin, O. & Darst, S.A. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296, 1285–1290 (2002).
Gusarov, I. & Nudler, E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell 107, 437–449 (2001).
Legault, P., Li, J., Mogridge, J., Kay, L.E. & Greenblatt, J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell 93, 289–299 (1998).
Scharpf, M. et al. Antitermination in bacteriophage lambda. The structure of the N36 peptide-boxB RNA complex. Eur. J. Biochem. 267, 2397–2408 (2000).
Su, L. et al. RNA recognition by a bent alpha-helix regulates transcriptional antitermination in phage lambda. Biochemistry 36, 12722–12732 (1997).
Jucker, F.M., Heus, H.A., Yip, P.F., Moors, E.H. & Pardi, A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 264, 968–980 (1996).
Heus, H.A. & Pardi, A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253, 191–194 (1991).
Franklin, N.C. Clustered arginine residues of bacteriophage lambda N protein are essential to antitermination of transcription, but their locale cannot compensate for boxB loop defects. J. Mol. Biol. 231, 343–360 (1993).
Barrick, J.E., Takahashi, T.T., Ren, J., Xia, T. & Roberts, R.W. Large libraries reveal diverse solutions to an RNA recognition problem. Proc. Natl. Acad. Sci. USA 98, 12374–12378 (2001).
Barrick, J.E., Takahashi, T.T., Balakin, A. & Roberts, R.W. Selection of RNA-binding peptides using mRNA-peptide fusions. Methods 23, 287–293 (2001).
Barrick, J.E. & Roberts, R.W. Sequence analysis of an artificial family of RNA-binding peptides. Protein Sci. 11, 2688–2696 (2002).
Xia, T. et al. The RNA-protein complex: direct probing of the interfacial recognition dynamics and its correlation with biological functions. Proc. Natl. Acad. Sci. USA 100, 8119–8123 (2003).
Cai, Z. et al. Solution structure of P22 transcriptional antitermination N peptide–boxB RNA complex. Nat. Struct. Biol. 5, 203–212 (1998).
Harada, K., Martin, S.S. & Frankel, A.D. Selection of RNA-binding peptides in vivo. Nature 380, 175–179 (1996).
Doelling, J.H. & Franklin, N.C. Effects of all single base substitutions in the loop of boxB on antitermination of transcription by bacteriophage lambda's N protein. Nucleic Acids Res. 17, 5565–5577 (1989).
Das, A. et al. Components of multiprotein-RNA complex that controls transcription elongation in Escherichia coli phage lambda. Methods Enzymol. 274, 374–402 (1996).
Friedman, D.I. In The Bacteriophages vol. 2 (ed. Calendar, R.) 263–318 (Plenum, New York, 1988).
Whalen, W., Ghosh, B. & Das, A. NusA protein is necessary and sufficient in vitro for phage lambda N gene product to suppress a rho-independent terminator placed downstream of nutL. Proc. Natl. Acad. Sci. USA 85, 2494–2498 (1988).
Gill, S.C., Weitzel, S.E. & von Hippel, P.H. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J. Mol. Biol. 220, 307–324 (1991).
Mason, S.W., Li, J. & Greenblatt, J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J. Biol. Chem. 267, 19418–19426 (1992).
Rees, W.A., Weitzel, S.E., Yager, T.D., Das, A. & von Hippel, P.H. Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc. Natl. Acad. Sci. USA 93, 342–346 (1996).
Chattopadhyay, S., Garcia-Mena, J., DeVito, J., Wolska, K. & Das, A. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage lambda. Proc. Natl. Acad. Sci. USA 92, 4061–4065 (1995).
Mogridge, J. et al. Independent ligand-induced folding of the RNA-binding domain and two functionally distinct antitermination regions in the phage lambda N protein. Mol. Cell. 1, 265–275 (1998).
Barik, S., Ghosh, B., Whalen, W., Lazinski, D. & Das, A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell 50, 885–899 (1987).
Mason, S.W. & Greenblatt, J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 5, 1504–1512 (1991).
Toulokhonov, I., Artsimovitch, I. & Landick, R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science 292, 730–733 (2001).
Marciniak, R.A., Calnan, B.J., Frankel, A.D. & Sharp, P.A. HIV-1 Tat protein trans-activates transcription in vitro. Cell 63, 791–802 (1990).
Richter, S., Ping, Y.H. & Rana, T.M. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 99, 7928–7933 (2002).
Steinmetz, E.J. & Brow, D.A. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16, 6993–7003 (1996).
Steinmetz, E.J. & Brow, D.A. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc. Natl. Acad. Sci. USA 95, 6699–6704 (1998).
Steinmetz, E.J., Conrad, N.K., Brow, D.A. & Corden, J.L. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413, 327–331 (2001).
Austin, R.J., Xia, T., Ren, J., Takahashi, T.T. & Roberts, R.W. Designed arginine-rich RNA-binding peptides with picomolar affinity. J. Am. Chem. Soc. 124, 10966–10967 (2002).
Milligan, J.F., Groebe, D.R., Witherell, G.W. & Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 (1987).
Rees, W.A., Weitzel, S.E., Das, A. & von Hippel, P.H. Regulation of the elongation-termination decision at intrinsic terminators by antitermination protein N of phage lambda. J. Mol. Biol. 273, 797–813 (1997).
Peled-Zehavi, H., Smith, C.A., Harada, K. & Frankel, A.D. Screening RNA-binding libraries by transcriptional antitermination in bacteria. Methods Enzymol. 318, 297–308 (2000).
Acknowledgements
We thank N. Franklin for the gifts of the two-plasmid N expressor–β-galactosidase reporter constructs, plasmid pMS7 and N–tester strain, P. von Hippel for the gift of pET-N1 plasmid and N protein, P. Legault for sharing the coordinates of the λ phage N peptide–boxB RNA complex and J.H. Richards, C.S. Parker and members of the Roberts laboratory for helpful comments on the manuscript. This work was supported by grants from US National Science Foundation (NSF) and National Institutes of Health (NIH) to R.W.R. R.W.R. is an Alfred P. Sloan research fellow, A.F. is an ACS postdoctoral fellow and T.T.T. was supported by an NIH training grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Xia, T., Frankel, A., Takahashi, T. et al. Context and conformation dictate function of a transcription antitermination switch. Nat Struct Mol Biol 10, 812–819 (2003). https://doi.org/10.1038/nsb983
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb983