Key Points
-
Alzheimer's disease is characterized by histological changes in the brain, including extracellular amyloid plaques that are composed of the amyloid-β peptide (Aβ), and intracellular neurofibrillary tangles that are composed of the microtubule associated protein TAU. The analysis of genes associated with inherited risk of Alzheimer's disease has indicated that increased production or decreased removal of Aβ is a key early event in the cascade that leads to these changes.
-
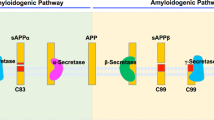
Aβ is derived from the amyloid precursor protein (APP), which undergoes a series of proteolytic cleavage events. The first, α-secretase event is mediated by members of the ADAM metalloproteinase family, and cleaves APP through the centre of the Aβ domain. This cleavage therefore precludes the formation of Aβ. In most cells, this is the predominant pathway for APP processing.
-
The other pathway involves two sequential cleavages of APP. The first, at the beginning of the Aβ-peptide domain, involves a membrane-bound aspartyl protease called β-secretase or BACE (β-site APP-cleaving enzyme). The second cleavage event, which clips the resulting β-stub in the centre of the transmembrane domain to generate the Aβ peptide and one or more carboxy-terminal fragments, is termed γ-secretase cleavage. The proteolytic cleavage of type 1 transmembrane proteins, such as APP, Notch, ErbB4 and Ire1p, within their hydrophobic transmembrane domains represents a novel form of regulated intramembranous proteolysis similar to that which cleaves SREBP (sterol-regulatory-element-binding protein) during the regulation of sterol metabolism.
-
Biochemical studies in cells in which the presenilin (PS) genes have been knocked out reveal that PS1 is required for and that PS2 facilitates this γ-secretase activity. In the absence of PS1, γ-secretase cleavage is significantly reduced, and in a double-knockout cell it is inhibited completely.
-
γ-Secretase cleavage of APP and the analogous S3-site cleavage of Notch generate a series of amino- and carboxy-terminal proteolytic products. Although all of the cleavage events involved in the generation of these products are presenilin-dependent, these cleavages of APP and Notch might be mediated by separate but similar pathways. The single known end-product of presenilin-dependent cleavage of Notch is the carboxy-terminal Notch intracellular domain, which derives from cleavage of Notch after residue 1743. This process is heavily dependent on the amino-acid sequence of Notch near the cleavage site. By contrast, cleavage of APP generates a series of amino-terminal fragments, including Aβ40, Aβ42 and rarer species, as well as a series of carboxy-terminal fragments, and is not dependent on the sequence of APP near these cleavage sites.
-
Some evidence has suggested that the presenilins might be either the catalytic site or a component of a multimeric γ-secretase enzyme complex: PS1 has weak sequence similarities to bacterial peptidases; transition-state analogue inhibitors of γ-secretase can be chemically cross-linked to PS1; mutation of two conserved aspartyl residues within transmembrane domains (which could be putative active-site residues of an aspartyl protease) inhibits γ-secretase cleavage; and PS1 and γ-secretase catalytic activities co-elute in purification studies.
-
But several observations do not fit easily with this hypothesis. First, there are subtle differences between the cleavage sites and sequence requirements for cleavage of APP and Notch during presenilin-mediated γ-like cleavage, so the catalytic activities might be carried out by other (possibly substrate-specific) components of the presenilin complex or by a distinct entity. In addition, mutation of both of the aspartyl residues does not completely suppress Aβ production, although it does cause accumulation of the substrates for γ-secretase cleavage (raising the possibility that PS1 is involved in trafficking substrates to or adapting substrates for the γ-secretase). Finally, although some studies using overexpressed presenilin transgenes suggest otherwise, most presenilin protein seems to be located within intracellular membranes. By contrast, most γ-like cleavage of Notch and APP seems to occur close to or after the plasmalemma (the 'spatial paradox').
Abstract
Investigations into the proteolytic processing of amyloid precursor protein (APP) have provided insights into both the pathogenesis of Alzheimer's disease and an unusual form of regulated proteolytic processing within the membrane-spanning domains of several proteins, including APP, Notch and ErbB4. Some of the enzymes responsible for α- and β-secretase cleavage have been identified, and these seem to be conventional proteolytic events. However, the molecular events that are involved in γ-secretase cleavage within the transmembrane domain of these proteins are much more complex. The presenilins and nicastrin are required for this process, but the role of the presenilins remains unclear. Although some data support the idea that the presenilins are in fact the active site of γ-secretase, other data indicate that they might have a more indirect role — for example, in transporting substrates to the correct subcellular compartments for γ-secretase cleavage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Price, D. L. & Sisodia, S. S. Mutant genes in familial Alzheimer's disease and transgenic models. Annu. Rev. Neurosci. 21, 479–505 (1998).
Schenk, D. et al. Immunization with Aβ attenuates Alzheimer's disease-like pathology in the PDAPP mouse. Nature 400, 173–177 (1999).
Bard, F. et al. Peripherally administered antibodies against amyloid β-peptide enter the central nervous system and reduce pathology in a mouse model of Alzheimer disease. Nature Med. 6, 916–919 (2000).
Janus, C. et al. Aβ-immunization reduces behavioural impairment and dense-cored plaques in a model of Alzheimer's disease. Nature 408, 979–982 (2000).
Morgan, D. et al. Aβ peptide vaccination prevents memory loss in an animal model of Alzheimer's disease. Nature 408, 982–985 (2000).References 2–5 describe the use of Aβ peptides as a vaccine to generate antibodies that facilitate the clearance of Alzheimer's-disease-like neuropathology and cognitive deficits in transgenic mouse models of Alzheimer's disease.
Fraser, P. E., Darabie, A. A. & McLaurin, J. A. Amyloid-β interactions with chondroitin sulfate-derived monosaccharides and disaccharides. Implications for drug development. J. Biol. Chem. 276, 6412–6419 (2001).
Soto, C. et al. β-Sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: implications for Alzheimer's therapy. Nature Med. 4, 822–826 (1998).
Kisilevsky, R. et al. Arresting amyloidosis in vivo using small-molecule anionic sulphonates or sulphates: implications for Alzheimer's disease. Nature Med. 1, 143–148 (1995).
Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398 (2000).
Kang, J. et al. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature 325, 733–736 (1987).
Goldgaber, D. et al. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer's disease. Science 235, 877–880 (1987).
Robakis, N. K. et al. in Disorders of the Developing Nervous System: Changing Views on their Origins, Diagnoses and Treatments (ed. Swann, J. W.) 183–193 (Alan R. Liss, New York, 1988).
Tanzi, R. E. et al. Amyloid β-protein gene: cDNA, mRNA distribution and genetic linkage near the Alzheimer locus. Science 235, 880–884 (1987).
Kitaguchi, N., Takahashi, Y., Tokushima, Y., Shiojiri, S. & Ito, H. Novel precursor of Alzheimer's disease amyloid protein shows protease inhibitory activity. Nature 331, 530–532 (1988).
Weidemann, A. et al. Identification, biogenesis, and localization of precursors of Alzheimer disease A4 amyloid protein. Cell 57, 115–126 (1989).
Haass, C., Koo, E. H., Mellon, A., Hung, A. Y. & Selkoe, D. J. Targeting of cell-surface β-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature 357, 500–503 (1992).
Sisodia, S. S. Secretion of the β-amyloid precursor protein. Ann. NY Acad. Sci. 674, 53–57 (1992).
Hung, A. Y. & Selkoe, D. J. Selective ectodomain phosphorylation and regulated cleavage of β-amyloid precursor protein. EMBO J. 13, 534–542 (1994).
Walter, J. et al. Ectodomain phosphorylation of β-amyloid precursor protein at two distinct cellular locations. J. Biol. Chem. 272, 1896–1903 (1997).
Zheng, H. et al. Mice deficient for the amyloid precursor protein gene. Ann. NY Acad. Sci. 777, 421–426 (1996).
Saitoh, T. et al. Secreted form of amyloid β-protein is involved in the growth regulation of fibroblasts. Cell 58, 615–622 (1989).
Milward, A. E. et al. The amyloid protein precursor of Alzheimer disease is a mediator of the effects of NGF on neurite outgrowth. Neuron 9, 129–137 (1992).
Nishimoto, I. et al. Alzheimer amyloid protein precursor complexes with brain GTP-binding protein Go. Nature 362, 75–79 (1993).
Kamal, A., Almenar-Queralt, A., LeBlanc, J. F., Roberts, E. A. & Goldstein, L. S. Kinesin-mediated axonal transport of a membrane compartment containing β-secretase and presenilin-1 requires APP. Nature 414, 643–648 (2001).
Cao, X. & Sudhof, T. C. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120 (2001).
Sastre, M. et al. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2, 835–841 (2001).
Cupers, P., Orlans, I., Craessaerts, K., Annaert, W. & De Strooper, B. The amyloid precursor protein (APP)-cytoplasmic fragment generated by γ-secretase is rapidly degraded but distributes partially in a nuclear fraction of neurones in culture. J. Neurochem. 78, 1168–1178 (2001).
Palmert, M. R. et al. The β-amyloid protein precursor of Alzheimer disease has soluble derivatives found in human brain and cerebrospinal fluid. Proc. Natl Acad. Sci. USA 86, 6338–6342 (1989).
Esch, F. S. et al. Cleavage of amyloid β-peptide during constitutive processing of its precursor. Science 248, 1122–1124 (1990).
Sisodia, S. S., Koo, E. H., Beyreuther, K., Unterbeck, A. & Price, D. Evidence that β-amyloid protein in Alzheimer's disease is not derived by normal processing. Science 248, 492–495 (1990).
Wang, R., Meschia, J. F., Cotter, R. J. & Sisodia, S. S. Secretion of the β/A4 amyloid precursor protein. Identification of a cleavage site in cultured mammalian cells. J. Biol. Chem. 266, 16960–16964 (1991).
Sahasrabudhe, S. R. et al. Enzymatic generation of the amino terminus of the β-amyloid peptide. J. Biol. Chem. 268, 16699–16705 (1993).
Buxbaum, J. D. et al. Evidence that tumor necrosis factor-α converting enzyme is involved in regulated α-secretase cleavage of the Alzheimer amyloid protein precursor. J. Biol. Chem. 273, 27765–27767 (1998).
Nitsch, R. M., Slack, B. E., Wurtman, R. J. & Growden, J. H. Release of Alzheimer amyloid precursor derivatives stimulated by activation of muscarinic acetylcholine receptors. Science 258, 304–307 (1992).
Xu, H., Greengard, P. & Gandy, S. Regulated formation of Golgi secretory vesicles containing Alzheimer β-amyloid precursor protein. J. Biol. Chem. 270, 23243–23245 (1995).
Parvathy, S., Karran, E. H., Turner, A. J. & Hooper, N. M. The secretases that cleave angiotensin converting enzyme and the amyloid precursor protein are distinct from tumour necrosis factor-α convertase. FEBS Lett. 431, 63–65 (1998).
Lammich, S. et al. Constitutive and regulated α-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl Acad. Sci. USA 96, 3922–3927 (1999).References 33–37 show that α-secretase cleavage is a physiological, regulated cleavage of APP, and that TACE and other members of the ADAM superfamily are good candidates for this enzymatic activity.
Vassar, R. et al. β-Secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741 (1999).
Hussain, I. et al. Identification of a novel aspartic protease (Asp2) as β-secretase. Mol. Cell. Neurosci. 14, 419–427 (1999).
Sinha, S. et al. Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature 402, 537–540 (1999).
Yan, R. et al. Membrane-anchored aspartyl protease with Alzheimer's disease β-secretase activity. Nature 402, 533–537 (1999).References 38–41 describe the cloning of β-secretase as a type 1 transmembrane aspartyl protease.
Shoji, M. et al. Production of the Alzheimer amyloid β protein by normal proteolytic processing. Science 258, 126–129 (1992).
Haass, C. et al. Amyloid β-peptide is produced by cultured cells during normal metabolism. Nature 359, 322–325 (1992).References 42 and 43 show that Aβ production is a physiological event in many cell types.
Jarrett, J. T. & Lansbury, P. T. Seeding one-dimensional crystallization of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell 73, 1055–1058 (1993).
Yankner, B. A., Duffy, L. K. & Kirschner, D. A. Neurotrophic and neurotoxic effects of amyloid β protein: reversal by tachykinin neuropeptides. Science 250, 279–282 (1990).
Pike, C. J., Burdick, D., Walencewicz, A. J., Glabe, C. G. & Cotman, C. W. Neurodegeneration induced by β-amyloid peptides in vitro: the role of peptide assembly state. J. Neurosci. 13, 1676–1678 (1993).
Lorenzo, A. & Yanker, B. A. β-Amyloid neurotoxicity requires fibril formation and is inhibited by Congo red. Proc. Natl Acad. Sci. USA 91, 12243–12247 (1994).
Koo, E. H. & Squazzo, S. L. Evidence that production and release of amyloid β-protein precursor involves the endocytic pathway. J. Biol. Chem. 269, 17386–17389 (1994).
Koo, E. H., Squazzo, S. L., Selkoe, D. J. & Koo, C. H. Trafficking of cell-surface amyloid β-protein precursor. I. Secretion, endocytosis and recycling as detected by labeled monoclonal antibody. J. Cell Sci. 109, 991–998 (1996).
Cook, D. G. et al. Alzheimer Aβ1–42 peptide is generated in the endoplasmic reticulum/intermediate compartment of NT2N cells. Nature Med. 3, 1021–1023 (1997).
Hartmann, T. et al. Distinct sites of intracellular production for Alzheimer's disease Aβ40/42 amyloid peptides. Nature Med. 3, 1016–1020 (1997).
Citron, M. et al. Evidence that the 42- and 40-amino-acid forms of amyloid β protein are generated from the β-amyloid precursor protein by different protease activities. Proc. Natl Acad. Sci. USA 93, 13170–13175 (1996).
Yasojima, K., Akiyama, H., McGeer, E. G. & McGeer, P. L. Reduced neprilysin in high plaque areas of Alzheimer brain: a possible relationship to deficient degradation of β-amyloid peptide. Neurosci. Lett. 297, 97–100 (2001).
Qiu, W. Q. et al. Insulin-degrading enzyme regulates extracellular levels of amyloid β-protein by degradation. J. Biol. Chem. 273, 32730–32738 (1998).
Chesneau, V., Vekrellis, K., Rosner, M. R. & Selkoe, D. J. Purified recombinant insulin-degrading enzyme degrades amyloid β-protein but does not promote its oligomerization. Biochem. J. 351, 509–516 (2000).
Goate, A. M. et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer disease. Nature 349, 704–706 (1991).
Sherrington, R. et al. Cloning of a gene bearing missense mutations in early onset familial Alzheimer's disease. Nature 375, 754–760 (1995).
Levy-Lahad, E. et al. A familial Alzheimer's disease locus on chromosome 1. Science 269, 970–973 (1995).
Rogaev, E. I. et al. Familial Alzheimer's disease in kindreds with missense mutations in a novel gene on chromosome 1 related to the Alzheimer's disease type 3 gene. Nature 376, 775–778 (1995).References 56–59 describe the discovery of disease-causing mutations in the APP, PS1 and PS2 genes.
Citron, M. et al. Mutation of the β-amyloid precursor protein in familial Alzheimer's disease increases β-protein production. Nature 360, 672–674 (1992).
Haass, C., Hung, A. Y., Selkoe, D. J. & Teplow, D. B. Mutations associated with a locus for familial Alzheimer's disease result in alternative processing of amyloid β-protein precursor. J. Biol. Chem. 269, 17741–17748 (1994).
Scheuner, D. et al. Secreted amyloid-β protein similar to that in the senile plaques of Alzheimer disease is increased in vivo by presenilin 1 and 2 and APP mutations linked to FAD. Nature Med. 2, 864–870 (1996).
Borchelt, D. R. et al. Familial Alzheimer's disease linked presenilin 1 variants elevate Aβ1–42/1–40 ratio in vitro and in vivo. Neuron 17, 1005–1013 (1996).
Duff, K. et al. Increased amyloid-β42(43) in brains of mice expressing mutant presenilin 1. Nature 383, 710–713 (1996).
Citron, M. et al. Mutant presenilins of Alzheimer's disease increase production of 42-residue amyloid β-protein in both transfected cells and transgenic mice. Nature Med. 3, 67–72 (1997).References 60–65 describe the discovery that disease-causing mutations in APP, PS1 and PS2 all alter the processing of APP and/or of Aβ itself, causing accumulation of neurotoxic derivatives.
L'Hernault, S. W. L. & Arduengo, P. M. Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J. Cell Biol. 119, 55–69 (1992).
Levitan, D. & Greenwald, I. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabditis elegans S182 Alzheimer's disease gene. Nature 377, 351–354 (1995).
Li, X. & Greenwald, I. HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signalling. Proc. Natl Acad. Sci. USA 94, 12204–12209 (1997).
Doan, A. et al. Protein topology of presenilin 1. Neuron 17, 1023–1030 (1996).
Li, X. & Greenwald, I. Membrane topology of the C. elegans SEL-12 presenilin. Neuron 17, 1015–1021 (1996).
De Strooper, B. et al. Phosphorylation, subcellular localization, and membrane orientation of the Alzheimer's disease-associated presenilins. J. Biol. Chem. 272, 3590–3598 (1997).
Thinakaran, G. et al. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron 17, 181–190 (1996).
Lee, M. K. et al. Hyperaccumulation of FAD-linked presenilin-1 variants in vivo. Nature Med. 3, 756–760 (1997).
Thinakaran, G. et al. Evidence that levels of presenilins (PS1 and PS2) are coordinately regulated by competition for limiting cellular factors. J. Biol. Chem. 272, 28415–28422 (1997).
Yu, G. et al. The presenilin 1 protein is a component of a high molecular weight intracellular complex that contains β-catenin. J. Biol. Chem. 273, 16470–16475 (1998).
Thinakaran, G. et al. Stable association of the presenilin derivatives and absence of presenilin interactions with APP. Neurobiol. Dis. 4, 438–453 (1998).
Capell, A. et al. The proteolytic fragments of the Alzheimer's disease associated presenilin-1 form heterodimers and occur as a 100–150 kDa molecular mass complex. J. Biol. Chem. 273, 3205–3211 (1998).
Li, Y.-M. et al. Presenilin 1 is linked with γ-secretase activity in the detergent solubilized state. Proc. Natl Acad. Sci. USA 97, 6138–6143 (2000).References 75–78 describe the discovery that PS1 is part of a multimeric, high-molecular-weight protein complex.
Walter, J. et al. The Alzheimer's disease associated presenilins are differentially phosphorylated proteins located predominantly within the endoplasmic reticulum. Mol. Med. 2, 673–691 (1996).
Annaert, W. G. et al. Presenilin 1 controls γ-secretase processing of amyloid precursor protein in pre-Golgi compartments of hippocampal neurons. J. Cell Biol. 147, 277–294 (1999).
Cupers, P. et al. The discrepancy between presenilin subcellular localization and γ-secretase processing of amyloid precursor protein. J. Cell Biol. 154, 731–740 (2001).
Ray, W. J. et al. Cell surface presenilin 1 participates in the γ-secretase-like proteolysis of Notch. J. Biol. Chem. 274, 36801–36807 (1999).
Baki, L. et al. Presenilin-1 binds cytoplasmic epithelial cadherin, inhibits cadherin/p120 association, and regulates stability and function of the cadherin/catenin adhesion complex. Proc. Natl Acad. Sci. USA 98, 2381–2386 (2001).
Georgakopoulos, A. et al. Presenilin-1 forms complexes with the cadherin/catenin cell–cell adhesion system and is recruited to intercellular and synaptic contacts. Mol. Cell 4, 893–902 (1999).
Chung, H.-M. & Struhl, G. Nicastrin is required for Presenilin-mediated transmembrane cleavage in Drosophila. Nature Cell Biol. 3, 1129–1132 (2001).
Zhou, J. et al. Presenilin 1 interacts with a novel member of the armadillo family. Neuroreport 8, 2085–2090 (1997).
Levesque, G. et al. Presenilins interact with armadillo proteins including neural specific plakophilin related protein and β-catenin. J. Neurochem. 72, 999–1008 (1999).
Yu, G. et al. Nicastrin modulates presenilin-mediated notch/glp-1 and βAPP processing. Nature 407, 48–54 (2000).Describes the discovery of a protein that is a component of the presenilin complex.
Saura, C. A. et al. The nonconserved hydrophilic loop domain of presenilin (PS) is not required for PS endoproteolysis or enhanced Aβ42 production mediated by familial early onset Alzheimer's disease-linked PS variants. J. Biol. Chem. 275, 17136–17142 (2000).
Ho, C. et al. δ-Catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J. Comp. Neurol. 420, 261–276 (2000).
Lu, Q. et al. δ-Catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 144, 519–532 (1999).
Goutte, C., Hepler, W., Mickey, K. M. & Priess, J. R. aph-2 encodes a novel extracellular protein required for GLP-1-mediated signaling. Development 127, 2481–2492 (2000).
Levitan, D., Yu, G., St George-Hyslop, P. & Goutte, C. APH-2/Nicastrin functions in LIN-12/Notch signalling in the C. elegans somatic gonad. Dev. Biol. 240, 654–661 (2001).
Lopez-Schier, H. & St Johnston, D. S. Drosophila Nicastrin is essential for the intramembranous cleavage of Notch. Dev. Cell 2, 79–89 (2002).
Hu, Y., Ye, Y. & Fortini, M. E. Nicastrin is required for γ-secretase cleavage of the Drosophila Notch receptor. Dev. Cell 2, 69–78 (2002).
Chen, F. et al. Nicastrin binds to membrane-tethered Notch. Nature Cell Biol. 3, 751–754 (2001).
De Strooper, B. et al. Deficiency of presenilin 1 inhibits the normal cleavage of amyloid precursor protein. Nature 391, 387–390 (1998).
Naruse, S. et al. Effect of PS1 deficiency on membrane protein trafficking in neurons. Neuron 21, 1213–1221 (1998).
Chen, F. et al. Proteolytic derivative of amyloid precursor protein accumulate in restricted and unpredicted intracellular compartments in the absence of functional presenilin 1 expression. J. Biol. Chem. 275, 36794–36802 (2000).
Herreman, A. et al. Total inactivation of γ-secretase activity in presenilin-deficient embryonic stem cells. Nature Cell Biol. 2, 461–462 (2000).
Zhang, Z. et al. Presenilins are required for γ-secretase cleavage of β-APP and transmembrane cleavage of Notch-1. Nature Cell Biol. 2, 463–465 (2000).References 97–101, 108–113 and 115–117 describe the discovery that PS1 (and the other components of the multimeric PS1 complex) are required for the cleavage of APP, ErbB4 and Notch within their transmembrane domains.
Brou, C. et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 (2000).
Mumm, J. S. et al. A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 (2000).
Sisodia, S. S. β-Amyloid precursor protein cleavage by a membrane-bound protease. Proc. Natl Acad. Sci. USA 89, 6075–6079 (1992).
Schroeter, E. H., Kisslinger, J. A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
Weinmaster, G. Notch signal transduction: a real rip and more. Curr. Opin. Genet. Dev. 10, 363–369 (2000).
Chan, Y. M. & Jan, Y. N. Presenilins, processing of β-amyloid precursor protein, and Notch signaling. Neuron 23, 201–204 (1999).
Wong, P. C. et al. Presenilin 1 is required for Notch and Dll1 expression in the paraxial mesoderm. Nature 387, 288–292 (1997).
Shen, J. et al. Skeletal and CNS defects in presenilin-1 deficient mice. Cell 89, 629–639 (1997).
Donoviel, D. et al. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13, 2801–2810 (1999).
Herreman, A. et al. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc. Natl Acad. Sci. USA 96, 11872–11877 (1999).
De Strooper, B. et al. A presenilin dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Ni, C. Y., Murphy, M. P., Golde, T. E. & Carpenter, G. γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294, 2179–2181 (2001).
Fergani, A., Yu, G., St George-Hyslop, P. & Checler, F. Wild-type and mutated nicastrins do not display aminopeptidase M- and B-like activities. Biochem. Biophys. Res. Commun. 289, 678–680 (2001).
Wolfe, M. S. et al. Two transmembrane aspartates in presenilin 1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 (1999).
Li, Y. M. et al. Photoactivated γ-secretase inhibitors directed to the active site covalently label presenilin 1. Nature 405, 689–694 (2000).
Esler, W. P. et al. Transition-state analogue inhibitors of γ-secretase bind directly to presenilin-1. Nature Cell Biol. 2, 428–434 (2000).
Steiner, H. et al. Glycine 384 is required for presenilin-1 function and is conserved in bacterial polytopic aspartyl proteases. Nature Cell Biol. 2, 848–851 (2000).
Huppert, S. S. et al. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature 405, 966–970 (2000).
Lichtenthaler, S. F., Ida, N., Multhaup, G., Masters, C. L. & Beyreuther, K. Mutations in the transmembrane domain of APP altering γ-secretase specificity. Biochemistry 36, 15396–15403 (1997).
Yu, C. et al. Characterization of a presenilin-mediated amyloid precursor protein carboxyl-terminal fragment γ. Evidence for distinct mechanisms involved in γ-secretase processing of the APP and Notch1 transmembrane domains. J. Biol. Chem. 276, 43756–43760 (2001).
Gu, Y. et al. Distinct intramembrane cleavage of the β-amyloid precursor protein family resembling γ-secretase-like cleavage of Notch. J. Biol. Chem. 276, 35235–35238 (2001).
Kulic, L. et al. Separation of presenilin function in amyloid β-peptide generation and endoproteolysis of Notch. Proc. Natl Acad. Sci. USA 97, 5913–5918 (2000).
Petit, A. et al. New protease inhibitors prevent γ-secretase-mediated Aβ40/42 production without affecting Notch cleavage. Nature Cell Biol. 3, 507–511 (2001).
Kim, S. H. et al. Multiple effects of aspartate mutant presenilin 1 on the processing and trafficking of amyloid precursor protein. J. Biol. Chem. 276, 43343–43350 (2001).
Capell, A. et al. Presenilin-1 differentially facilitates endoproteolysis of the β-amyloid precursor protein and Notch. Nature Cell Biol. 2, 205–211 (2000).
Murphy, M. P. et al. Presenilin 1 regulates pharmacologically distinct γ-secretase activities. Implications for the role of presenilin in γ-secretase cleavage. J. Biol. Chem. 275, 26277–26284 (2000).
Tomita, T. et al. The first proline of PALP motif at the C terminus of presenilins is obligatory for stabilization, complex formation, and γ-secretase activities of presenilins. J. Biol. Chem. 276, 33273–33281 (2001).
Yu, G. et al. Mutation of conserved aspartates affects maturation of both aspartate mutant and endogenous presenilin 1 and presenilin 2 complexes. J. Biol. Chem. 275, 27348–27353 (2000).
Perez, R. G. et al. Mutagenesis identifies new signals for β-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Aβ42 J. Biol. Chem. 274, 18851–18856 (1999).
Arduengo, P. M., Appleberry, O. K., Chuang, P. & L'Hernault, S. W. The presenilin protein family member SPE-4 localizes to an ER/Golgi derived organelle and is required for proper cytoplasmic partitioning during Caenorhabditis elegans spermatogenesis. J. Cell Sci. 111, 3645–3654 (1998).
Annaert, W. G. et al. Interaction with telencephalin and the amyloid precursor protein predicts a ring structure for presenilins. Neuron 32, 579–589 (2001).
DeBose-Boyd, R. A. et al. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99, 703–712 (1999).
Cai, H. et al. BACE1 is the major β-secretase for generation of Aβ peptides by neurons. Nature Neurosci. 4, 233–234 (2001).
Wong, P. C., Price, D. L. & Cai, H. The brain's susceptibility to amyloid plaques. Science 293, 1434 (2001).
Hadland, B. K. et al. γ-Secretase inhibitors repress thymocyte development. Proc. Natl Acad. Sci. USA 98, 7487–7491 (2001).
St George-Hyslop, P. H. Piecing together Alzheimer's. Sci. Am. 283, 76–83 (2000).
Acknowledgements
We acknowledge support from the US Public Health Service, the Metropolitan Life Foundation, the Adler Foundation, the Alzheimer's Association, the American Health Assistance Foundation, the Canadian Institutes of Health Research, the Alzheimer Society of Ontario and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
Entrez Protein
FlyBase
LocusLink
OMIM
FURTHER INFORMATION
Encyclopedia of Life Sciences
Glossary
- TYPE 1 TRANSMEMBRANE RECEPTORS
-
Transmembrane proteins that traverse the membrane once, with the carboxyl terminus in the cytoplasm and the amino terminus oriented towards the lumen or extracellular space.
- PC12 CELLS
-
Neuron-like cells that are derived from a malignant neural crest tumour (a phaeochromocytoma).
- HOLOPROTEIN
-
The full-length, native polypeptide before proteolytic cleavage events that might occur during maturation.
- ARMADILLO-REPEAT PROTEINS
-
An extensive family of highly conserved, related proteins that contain a series of amino-acid repeat elements (called arm-repeats), and which have diverse cellular functions, including structural and signal-transduction roles. This family includes the α-, β- and γ-catenins.
- RNA INTERFERENCE
-
(RNAi). A method by which double-stranded RNA that is encoded on an exogenous vector can be used to interfere with normal RNA processing, causing rapid degradation of the endogenous RNA and thereby precluding translation. This provides a simple way of studying the effects of the absence of a gene product in simple organisms and in cells.
- DOMINANT NEGATIVE
-
Describes a mutant molecule that can form a heteromeric complex with the normal molecule, knocking out the activity of the entire complex.
- CSL PROTEIN
-
CBF1, Su(H) and LAG-1 proteins are a series of downstream transcriptional regulators that are involved in the nuclear transduction of Notch-receptor-mediated signalling events at the cell surface.
- DI-ASPARTYL PROTEASE
-
A putative protease in which the active catalytic site contains two aspartyl residues.
- TRANSITION-STATE ANALOGUE
-
A compound that is designed to resemble an intermediate, transition-state complex between a protease and its substrate. It is intended to inactivate the protease in a 'dead-end' complex.
- SCISSILE BOND
-
The inter-amino-acid bond that is cleaved during proteolysis.
Rights and permissions
About this article
Cite this article
Sisodia, S., St George-Hyslop, P. γ-Secretase, notch, Aβ and alzheimer's disease: Where do the presenilins fit in?. Nat Rev Neurosci 3, 281–290 (2002). https://doi.org/10.1038/nrn785
Issue Date:
DOI: https://doi.org/10.1038/nrn785
This article is cited by
-
Role of Necroptosis, a Regulated Cell Death, in Seizure and Epilepsy
Neurochemical Research (2024)
-
Prolonged culturing of iPSC-derived brain endothelial-like cells is associated with quiescence, downregulation of glycolysis, and resistance to disruption by an Alzheimer’s brain milieu
Fluids and Barriers of the CNS (2022)
-
SARS-CoV-2 Spike protein S2 subunit modulates γ-secretase and enhances amyloid-β production in COVID-19 neuropathy
Cell Discovery (2022)
-
Nanoscale organization of Nicastrin, the substrate receptor of the γ-secretase complex, as independent molecular domains
Molecular Brain (2021)
-
Mulberry Fruit Extract Alleviates Cognitive Impairment by Promoting the Clearance of Amyloid-β and Inhibiting Neuroinflammation in Alzheimer’s Disease Mice
Neurochemical Research (2020)