Key Points
-
New insights into the structures of glutamate receptor ion channels (iGluRs), combined with functional and biochemical data, can help us to understand how agonist binding triggers their activation and subsequent desensitization. Studies of related channels also provide valuable insights into the molecular basis of ion selectivity and transport.
-
iGluRs are important for fast excitatory neural signalling and for synaptic plasticity. They are constructed from subunits that include an amino-terminal domain (NTD), a ligand-binding domain (S1S2), three transmembrane domains, a P-loop domain and a carboxy-terminal domain.
-
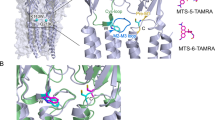
The ligand-binding domain, S1S2, can be expressed and studied in isolation. It consists of two lobes that form a cleft, which is open at rest. When it binds agonist, the cleft closes around the ligand through a conformational change involving the formation of hydrogen bonds between the agonist and S1S2. The mechanism of closure involves an open⇔closed equilibrium for the ligand-binding domain. The balance of equilibrium shifts from the open to the closed state when it interacts with agonist.
-
The ligand-binding site seems to possess several 'subsites' that participate in ligand binding. Not all of these subsites need to be occupied for agonist binding, providing the flexibility to bind various agents. Vibrational spectroscopy shows that electronegative ligand moieties interact with electronegative groups in the binding site.
-
It is not clear how the receptor subunits combine to form an ion channel. It is proposed that tetrameric channels form as a 'dimer of dimers', with initial dimerization being mediated by the NTDs, and the second dimerization of dimers requiring compatibility between the S2 and transmembrane domains.
-
Current models of channel activation and desensitization involve cleft closure in each subunit, pulling the transmembrane domains of the subunit away from the pore axis, and thus opening the channel. The channel then closes by slippage between subunits.
-
Glutamate receptors are permeable to both K+ and Na+, and in some cases to divalent cations. It seems likely that the channel opens wide enough to allow ions to pass through with their hydration shells. This contrasts with the K+ channels, in which ions are dehydrated at the entrance to the channel.
-
Many questions remain to be answered about the ligand binding, activation, ion permeation and desensitization of iGluRs. The packing and symmetry of the P-loop sequence are unclear, as are the assembly interactions of the subunits. As these details become better understood, they will help us to understand the kinetics and selectivity of glutamate-gated channels.
Abstract
As in the case of many ligand-gated ion channels, the biochemical and electrophysiological properties of the ionotropic glutamate receptors have been studied extensively. Nevertheless, we still do not understand the molecular mechanisms that harness the free energy of agonist binding, first to drive channel opening, and then to allow the channel to close (desensitize) even though agonist remains bound. Recent crystallographic analyses of the ligand-binding domains of these receptors have identified conformational changes associated with agonist binding, yielding a working hypothesis of channel function. This opens the way to determining how the domains and subunits are assembled into an oligomeric channel, how the domains are connected, how the channel is formed, and where it is located relative to the ligand-binding domains, all of which govern the processes of channel activation and desensitization.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hamill, O. P., Marty, A., Neher, E., Sakmann, B. & Sigworth, F. J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflügers Arch. 391, 85–100 (1981).
Sakmann, B. & Neher, E. (eds) Single Channel Recording (Plenum, New York, 1983).
Armstrong, N., Sun, Y., Chen, G.-Q. & Gouaux, E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature 395, 913–917 (1998). The first atomic-resolution structure of a glutamate receptor ligand-binding domain.
Armstrong, N. A. & Gouaux, E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181 (2000). Crystallographic structures of the iGluR ligand-binding domain in the apo state and in a complex with agonists and antagonists revealed cleft closure on agonist binding. The extent of closure correlates with the extent of channel activation by a given agonist, providing the first model of iGluR activation.
Mayer, M. L., Olson, R. & Gouaux, E. Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state. J. Mol. Biol. 311, 815–836 (2001). This study provides the first evidence of dimerization of the ligand-binding domain in solution. The crystallographic structure of the ligand-free prokaryotic iGluR ligand-binding domain revealed a closed cleft, indicating an open–closed equilibrium that is probably reflected in spontaneous activity of the channel.
Doyle, D. A. et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 (1998). The first crystallographic structure of an ion channel, revealing mechanisms by which ion selectivity is combined with high flux.
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001).
Unwin, N. The Croonian lecture 2000. Nicotinic acetylcholine receptor and the structural basis of fast synaptic transmission. Philos Trans R Soc Lond B Biol Sci 355, 1813–1829 (2000).
Brejc, K. et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276 (2001). The crystal structure of an acetylcholine-binding protein pentamer reveals the location of ligand-binding sites at the subunit interfaces.
Dingledine, R., Borges, K., Bowie, D. & Traynelis, S. F. The glutamate receptor ion channels. Pharmacol. Rev. 51, 7–61 (1999).
Hollmann, M. & Heinemann, S. Cloned glutamate receptors. Annu. Rev. Neurosci. 17, 31–108 (1994).
Hollmann, M., Maron, C. & Heinemann, S. N -glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron 13, 1331–1343 (1994).
O'Hara, P. J. et al. The ligand-binding domain of metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron 11, 41–52 (1993).
Choi, Y. B. & Lipton, S. A. Identification and mechanism of action of two histidine residues underlying high-affinity Zn2+ inhibition of the NMDA receptor. Neuron 23, 171–180 (1999).
Fayyazuddin, A., Villarroel, A., Le Goff, A., Lerma, J. & Neyton, J. Four residues of the extracellular N-terminal domain of the NR2A subunit control high-affinity Zn2+ binding to NMDA receptors. Neuron 25, 683–694 (2000).
Low, C. M., Zheng, F., Lyuboslavsky, P. & Traynelis, S. F. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-d-aspartate NR1/NR2A receptors. Proc. Natl Acad. Sci. USA 97, 11062–11067 (2000).
Paoletti, P. et al. Molecular organization of a zinc binding N-terminal modulatory domain in a NMDA receptor subunit. Neuron 28, 911–925 (2000).
Gallagher, M. J., Huang, H., Pritchett, D. B. & Lynch, D. R. Interactions between ifenprodil and the NR2B subunit of the N-methyl-d-aspartate receptor. J. Biol. Chem. 271, 9603–9611 (1996).
Gallagher, M. J., Huang, H., Grant, E. R. & Lynch, D. R. The NR2B-specific interactions of polyamines and protons with the N-methyl-d-aspartate receptor. J. Biol. Chem. 272, 24971–24979 (1997).
Brimecombe, J. C., Gallagher, M. J., Lynch, D. R. & Aizenman, E. An NR2B point mutation affecting haloperidol and CP101,606 sensitivity of single recombinant N-methyl-d-aspartate receptors. J. Pharmacol. Exp. Ther. 286, 627–634 (1998).
Gallagher, M. J., Huang, H. & Lynch, D. R. Modulation of the N-methyl-d-aspartate receptor by haloperidol: NR2B-specific interactions. J. Neurochem. 70, 2120–2128 (1998).
Shim, S. S., Grant, E. R., Singh, S., Gallagher, M. J. & Lynch, D. R. Actions of butyrophenones and other antipsychotic agents at NMDA receptors: relationship with clinical effects and structural considerations. Neurochem. Int. 34, 167–175 (1999).
Lynch, D. R. et al. Pharmacological characterization of interactions of RO 25-6981 with the NR2B (ɛ2) subunit. Eur. J. Pharmacol. 416, 185–195 (2001).
Masuko, T. et al. A regulatory domain (R1–R2) in the amino terminus of the N-methyl-d-aspartate receptor: effects of spermine, protons, and ifenprodil, and structural similarity to bacterial leucine/isoleucine/valine binding protein. Mol. Pharmacol. 55, 957–969 (1999).
Zheng, F. et al. Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nature Neurosci. 4, 894–901 (2001).
Krupp, J. J., Vissel, B., Heinemann, S. F. & Westbrook, G. L. N-terminal domains in the NR2 subunit control desensitization of NMDA receptors. Neuron 20, 317–327 (1998).
Villarroel, A., Regalado, M. P. & Lerma, J. Glycine-independent NMDA receptor desensitization — localization of structural determinants. Neuron 20, 329–339 (1998).
Kuusinen, A., Abele, R., Madden, D. R. & Keinänen, K. Oligomerization and ligand-binding properties of the ectodomain of the AMPA receptor subunit GluRD. J. Biol. Chem. 274, 28937–28943 (1999).
Leuschner, W. D. & Hoch, W. Subtype-specific assembly of α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunits is mediated by their N-terminal domains. J. Biol. Chem. 274, 16907–16916 (1999).
Ayalon, G. & Stern-Bach, Y. Functional assembly of AMPA and kainate receptors is mediated by several discrete protein–protein interactions. Neuron 31, 103–113 (2001). A detailed analysis of the intersubunit interactions that govern iGluR assembly indicates that the receptors are assembled as dimers of dimers.
Wada, K. et al. Sequence and expression of a frog brain complementary DNA encoding a kainate-binding protein. Nature 342, 684–689 (1989).
Gregor, P., Mano, I., Maoz, I., McKeown, M. & Teichberg, V. I. Molecular structure of the chick cerebellar kainate-binding subunit of a putative glutamate receptor. Nature 342, 689–692 (1989).
Wo, Z. G. & Oswald, R. E. Transmembrane topology of two kainate receptor subunits revealed by N-glycosylation. Proc. Natl Acad. Sci. USA 91, 7154–7158 (1994).
Nakanishi, N., Schneider, N. A. & Axel, R. A family of glutamate receptor genes: evidence for the formation of heteromultimeric receptors with distinct channel properties. Neuron 5, 569–581 (1990).
Hsiao, C. D., Sun, Y. J., Rose, J. & Wang, B. C. The crystal structure of glutamine-binding protein from Escherichia coli. J. Mol. Biol. 262, 225–242 (1996).
Wo, Z. G. & Oswald, R. E. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 18, 161–168 (1995).
Wood, M. W., VanDongen, H. M. & VanDongen, A. M. Structural conservation of ion conduction pathways in K channels and glutamate receptors. Proc. Natl Acad. Sci. USA 92, 4882–4886 (1995).
Verdoorn, T. A., Burnashev, N., Monyer, H., Seeburg, P. H. & Sakmann, B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science 252, 1715–1718 (1991).
Burnashev, N. et al. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science 257, 1415–1419 (1992).
Kuner, T., Wollmuth, L. P., Karlin, A., Seeburg, P. H. & Sakmann, B. Structure of the NMDA receptor channel M2 segment inferred from the accessibility of substituted cysteines. Neuron 17, 343–352 (1996).
Panchenko, V. A., Glasser, C. R. & Mayer, M. L. Structural similarities between glutamate receptor channels and K+ channels examined by scanning mutagenesis. J. Gen. Physiol. 117, 345–359 (2001).
Bolton, M. M., Blanpied, T. A. & Ehlers, M. D. Localization and stabilization of ionotropic glutamate receptors at synapses. Cell. Mol. Life Sci. 57, 1517–1525 (2000).
Gilbert, W. Why genes in pieces? Nature 271, 501 (1978).
Chen, G.-Q., Cui, C., Mayer, M. L. & Gouaux, E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature 402, 817–821 (1999). The identification of a prokaryotic iGluR provides molecular evidence that the iGluRs are derived from the fusion of a PBP with a primitive K+ channel.
Kuusinen, A., Arvola, M. & Keinänen, K. Molecular dissection of the agonist binding site of an AMPA receptor. EMBO J. 14, 6327–6332 (1995). The first demonstration of the ability to express functional iGluR ligand-binding domains as independent soluble proteins.
Chen, G.-Q. & Gouaux, E. Overexpression of a glutamate receptor (GluR2) ligand binding domain in Escherichia coli : application of a novel protein folding screen. Proc. Natl Acad. Sci. USA 94, 13431–13436 (1997).
Keinänen, K., Jouppila, A. & Kuusinen, A. Characterization of the kainate-binding domain of the glutamate receptor GluR-6 subunit. Biochem. J. 330, 1461–1467 (1998).
Ivanovic, A., Reiländer, H., Laube, B. & Kuhse, J. Expression and initial characterization of a soluble glycine binding domain of the N-methyl-d-aspartate receptor NR1 subunit. J. Biol. Chem. 273, 19933–19937 (1998).
Miyazaki, J., Nakanishi, S. & Jingami, H. Expression and characterization of a glycine-binding fragment of the N-methyl-d-aspartate receptor subunit NR1. Biochem. J. 340, 687–692 (1999).
Abele, R., Lampinen, M., Keinänen, K. & Madden, D. R. Disulfide bonding and cysteine accessibility in the α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor subunit GluRD: implications for redox modulation of glutamate receptors. J. Biol. Chem. 273, 25132–25138 (1998).
Rosenmund, C., Stern-Bach, Y. & Stevens, C. F. The tetrameric structure of a glutamate receptor channel. Science 280, 1596–1599 (1998). The analysis of gating transitions in iGluR under non-desensitizing conditions revealed incremental opening of the conducting pore, consistent with independent gating by four subunits assembled as a tetramer.
Oswald, R. E. & Changeux, J.-P. Crosslinking of α-bungarotoxin to the acetylcholine receptor from Torpedo marmorata by ultraviolet light irradiation. FEBS Lett. 139, 225–229 (1982).
Mao, B., Pear, M. R., McCammon, J. A. & Quiocho, F. A. Hinge-bending in l-arabinose-binding protein. The 'Venus's-flytrap' model. J. Biol. Chem. 257, 1131–1133 (1982).
Sharff, A. J., Rodseth, L. E., Spurlino, J. C. & Quiocho, F. A. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry 31, 10657–10663 (1992).
Oh, B.-H. et al. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J. Biol. Chem. 268, 11348–11355 (1993).
Oh, B.-H., Ferro-Luzzi Ames, G. & Kim, S.-H. Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein. J. Biol. Chem. 269, 26323–26330 (1994).
Schumacher, M. A., Choi, K. Y., Zalkin, H. & Brennan, R. G. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by α helices. Science 266, 763–770 (1994).
Schumacher, M. A., Choi, K. Y., Lu, F., Zalkin, H. & Brennan, R. G. Mechanism of corepressor-mediated specific DNA binding by the purine repressor. Cell 83, 147–155 (1995).
Lewis, M. et al. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271, 1247–1254 (1996).
Mowbray, S. L. & Björkman, A.J. Conformational changes of ribose-binding protein and two related repressors are tailored to fit the functional need. J. Mol. Biol. 294, 487–499 (1999).
Abele, R., Svergun, D., Keinänen, K., Koch, M. H. J. & Madden, D. R. A molecular envelope of the ligand-binding domain of a glutamate receptor in the presence and absence of agonist. Biochemistry 38, 10949–10957 (1999).
Patneau, D. K., Vyklicky, L. Jr & Mayer, M. L. Hippocampal neurons exhibit cyclothiazide-sensitive rapidly desensitizing responses to kainate. J. Neurosci. 13, 3496–3509 (1993).
Abele, R., Keinänen, K. & Madden, D. R. Agonist-induced isomerization in a glutamate receptor ligand-binding domain: a kinetic and mutagenetic analysis. J. Biol. Chem. 275, 21355–21363 (2000). The kinetics of iGluR agonist binding reveal an intermediate, fast docking step that precedes the slower cleft closure; key side chains are identified in both steps.
Kunishima, N. et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407, 971–977 (2000). The first crystallographic analysis of the ligand-binding domain of a metabotropic glutamate receptor shows open and closed conformations for both the empty and agonist-bound forms.
Wolf, A., Shaw, E. W., Nikaido, K. & Ferro-Luzzi Ames, G. The histidine-binding protein undergoes conformational changes in the absence of ligand as analyzed with conformation-specific monoclonal antibodies. J. Biol. Chem. 269, 23051–23058 (1994).
Careaga, C. L., Sutherland, J., Sabeti, J. & Falke, J. J. Large amplitude twisting motions of an interdomain hinge — a disulfide trapping study of the galactose–glucose binding protein. Biochemistry 34, 3048–3055 (1995).
Walmsley, A. R., Shaw, J. G. & Kelly, D. J. Perturbation of the equilibrium between open and closed conformations of the periplasmic C4-dicarboxylate binding protein from Rhodobacter capsulatus. Biochemistry 31, 11175–11181 (1992).
Flocco, M. M. & Mowbray, S. L. The 1.9 Å X-ray structure of a closed unliganded form of the periplasmic glucose/galactose receptor from Salmonella typhimurium. J. Biol. Chem. 269, 8931–8936 (1994).
Sack, J. S., Saper, M. A. & Quiocho, F. A. Periplasmic binding protein structure and function. Refined X-ray structures of the leucine/isoleucine/valine-binding protein and its complex with leucine. J. Mol. Biol. 206, 171–191 (1989).
Duan, X. Q., Hall, J. A., Nikaido, H. & Quiocho, F. A. Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding. J. Mol. Biol. 306, 1115–1126 (2001).
Vermersch, P. S., Lemon, D. D., Tesmer, J. J. G. & Quiocho, F. A. Sugar-binding and crystallographic studies of an arabinose-binding protein mutant (Met108Leu) that exhibits enhanced affinity and altered specificity. Biochemistry 30, 6861–6866 (1991).
Ledvina, P. S., Tsai, A.-L., Wang, Z., Koehl, E. & Quiocho, F. A. Dominant role of local dipolar interactions in phosphate binding to a receptor cleft with an electronegative charge surface: equilibrium, kinetic, and crystallographic studies. Protein Sci. 7, 2550–2559 (1998).
Walmsley, A. R., Shaw, J. G. & Kelly, D. J. The mechanism of ligand binding to the periplasmic C4-dicarboxylate binding protein (DctP) from Rhodobacter capsulatus. J. Biol. Chem. 267, 8064–8072 (1992).
Turecek, R., Vlachova, V. & Vyklicky, L. Spontaneous openings of NMDA receptor channels in cultured rat hippocampal neurons. Eur. J. Neurosci. 9, 1999–2008 (1997).
Zuo, J. et al. Neurodegeneration in Lurcher mice caused by mutation in δ2 glutamate receptor gene. Nature 388, 769–773 (1997).
Zheng, Y., Brockie, P. J., Mellem, J. E., Madsen, D. M. & Maricq, A. V. Neuronal control of locomotion in C. elegans is modified by a dominant mutation in the GLR-1 ionotropic glutamate receptor. Neuron 24, 347–361 (1999).
Kohda, K., Wang, Y. & Yuzaki, M. Mutation of a glutamate receptor motif reveals its role in gating and δ2 receptor channel properties. Nature Neurosci. 3, 315–322 (2000).
Schwarz, M. K. et al. Dominance of the lurcher mutation in heteromeric kainate and AMPA receptor channels. Eur. J. Neurosci. 14, 861–868 (2001).
Changeux, J.-P. & Edelstein, S. J. Allosteric mechanisms in normal and pathological nicotinic acetylcholine receptors. Curr. Opin. Neurobiol. 11, 369–377 (2001).
Paas, Y. The macro- and microarchitectures of the ligand-binding domain of glutamate receptors. Trends Neurosci. 21, 117–125 (1998).
Changeux, J.-P. & Edelstein, S. J. Allosteric receptors after 30 years. Neuron 21, 959–980 (1998).
Grosman, C., Zhou, M. & Auerbach, A. Mapping the conformational wave of acetylcholine receptor channel gating. Nature 403, 773–776 (2000).
Monod, J., Wyman, J. & Changeux, J.-P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 (1965).
Taverna, F. et al. The Lurcher mutation of an α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunit enhances potency of glutamate and converts an antagonist to an agonist. J. Biol. Chem. 275, 8475–8479 (2000).
Chapman, M. L., VanDongen, H. M. & VanDongen, A. M. Activation-dependent subconductance levels in the drk1 K channel suggest a subunit basis for ion permeation and gating. Biophys. J. 72, 708–719 (1997).
Zheng, J. & Sigworth, F. J. Intermediate conductances during deactivation of heteromultimeric Shaker potassium channels. J. Gen. Physiol. 112, 457–474 (1998).
Zheng, J. & Sigworth, F. J. Selectivity changes during activation of mutant Shaker potassium channels. J. Gen. Physiol. 110, 101–117 (1997).
Luecke, H. & Quiocho, F. A. High specificity of a phosphate transport protein determined by hydrogen bonds. Nature 347, 402–406 (1990).
Bräuner-Osborne, H., Egebjerg, J., Nielsen, E. O., Madsen, U. & Krogsgaard-Larsen, P. Ligands for glutamate receptors: design and therapeutic prospects. J. Med. Chem. 43, 2609–2645 (2000).
Jayaraman, V., Keesey, R. & Madden, D. R. Ligand–protein interactions in the glutamate receptor. Biochemistry 39, 8693–8697 (2000).
Madden, D. R., Thiran, S., Zimmermann, H., Romm, J. & Jayaraman, V. Stereochemistry of quinoxaline antagonist binding to a glutamate receptor investigated by Fourier transform infrared spectroscopy. J. Biol. Chem. 276, 37821–37826 (2001).
Laube, B., Kuhse, J. & Betz, H. Evidence for a tetrameric structure of recombinant NMDA receptors. J. Neurosci. 18, 2954–2961 (1998).
Mano, I. & Teichberg, V. I. A tetrameric subunit stoichiometry for a glutamate receptor-channel complex. Neuroreport 9, 327–331 (1998).
Safferling, M. et al. First images of a glutamate receptor ion channel: oligomeric state and molecular dimensions of GluRB homomers. Biochemistry 40, 13948–13953 (2001). This paper provides the first electron-microscopic views of an intact iGluR, showing it to be an elongated molecule of dimensions consistent with a dimer-of-dimers assembly.
Partin, K. M. Domain interactions regulating AMPA receptor desensitization. J. Neurosci. 21, 1939–1948 (2001).
Liu, D. T., Tibbs, G. R., Paoletti, P. & Siegelbaum, S. A. Constraining ligand-binding site stoichiometry suggests that a cyclic nucleotide-gated channel is composed of two functional dimers. Neuron 21, 235–248 (1998).
Perozo, E., Cortes, D. M. & Cuello, L. G. Structural rearrangements underlying K+-channel activation gating. Science 285, 73–78 (1999). Electron paramagnetic resonance spectroscopic analysis allowed the identification of conformational changes associated with gating of a K+ channel.
Patneau, D. K. & Mayer, M. L. Kinetic analysis of interactions between kainate and AMPA: evidence for activation of a single receptor in mouse hippocampal neurons. Neuron 6, 785–798 (1991).
Benveniste, M. & Mayer, M. L. Trapping of glutamate and glycine during open channel block of rat hippocampal neuron NMDA receptors by 9-aminoacridine. J. Physiol. (Lond.) 483, 367–384 (1995).
Lampinen, M., Pentikäinen, O., Johnson, M. S. & Keinänen, K. AMPA receptors and bacterial periplasmic amino acid-binding proteins share the ionic mechanism of ligand recognition. EMBO J. 17, 4704–4711 (1998).
Morais-Cabral, J. H., Zhou, Y. & MacKinnon, R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature 414, 37–42 (2001).
Lu, Z., Klem, A. M. & Ramu, Y. Ion conduction pore is conserved among potassium channels. Nature 413, 809–813 (2001).
Unwin, N. Acetylcholine receptor channel imaged in the open state. Nature 373, 37–43 (1995). Electron-microscopic analysis of different conformational states of the nAChR reveals a putative gating transition in the transmembrane domain M2.
Maeno, T., Edwards, C. & Anraku, M. Permeability of the endplate membrane activated by acetylcholine to some organic cations. J. Neurobiol. 8, 173–184 (1977).
Dwyer, T. M., Adams, D. J. & Hille, B. The permeability of the endplate channel to organic cations in frog muscle. J. Gen. Physiol. 75, 469–492 (1980).
Cohen, B. N., Labarca, C., Davidson, N. & Lester, H. A. Mutations in M2 alter the selectivity of the mouse nicotinic acetylcholine receptor for organic and alkali metal cations. J. Gen. Physiol. 100, 373–400 (1992).
Burnashev, N., Villarroel, A. & Sakmann, B. Dimensions and ion selectivity of recombinant AMPA and kainate receptor channels and their dependence on Q/R site residues. J. Physiol. (Lond.) 496, 165–173 (1996).
Villaroel, A., Burnashev, N. & Sakmann, B. Dimensions of the narrow portion of a recombinant NMDA receptor channel. Biophys. J. 68, 866–875 (1995).
Kuner, T., Beck, C., Sakmann, B. & Seeburg, P. H. Channel-lining residues of the AMPA receptor M2 segment: structural environment of the Q/R site and identification of the selectivity filter. J. Neurosci. 21, 4162–4172 (2001).
McCleskey, E. W. & Almers, W. The Ca channel in skeletal muscle is a large pore. Proc. Natl Acad. Sci. USA 82, 7149–7153 (1985).
Heginbotham, L., Abramson, T. & MacKinnon, R. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science 258, 1152–1155 (1992).
Heginbotham, L., Lu, Z., Abramson, T. & MacKinnon, R. Mutations in the K+ channel signature sequence. Biophys. J. 66, 1061–1067 (1994).
Sommer, B. et al. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science 249, 1580–1585 (1990).
Hollmann, M., O'Shea-Greenfield, A., Rodgers, S. W. & Heinemann, S. Cloning by functional expression of a member of the glutamate receptor family. Nature 342, 643–648 (1989).
Keinänen, K. et al. A family of AMPA-selective glutamate receptors. Science 249, 556–560 (1990).
Boulter, J. et al. Molecular cloning and functional expression of glutamate receptor subunit genes. Science 249, 1033–1037 (1990).
Rose, C. R. & Konnerth, A. Self-regulating synapses. Nature 405, 413–415 (2000).
Bettler, B. et al. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron 5, 583–595 (1990).
Egebjerg, J., Bettler, B., Hermans-Borgmeyer, I. & Heinemann, S. Cloning of a cDNA for a glutamate receptor subunit activated by kainate but not AMPA. Nature 351, 745–748 (1991).
Werner, P., Voigt, M., Keinänen, K., Wisden, W. & Seeburg, P. H. Cloning of a putative high-affinity kainate receptor expressed predominantly in hippocampal CA3 cells. Nature 351, 742–744 (1991).
Bettler, B. et al. Cloning of a putative glutamate receptor: a low affinity kainate-binding subunit. Neuron 8, 257–265 (1992).
Herb, A. et al. The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron 8, 775–785 (1992).
Lerma, J., Paternain, A. V., Rodriguez-Moreno, A. & Lopez-Garcia, J. C. Molecular physiology of kainate receptors. Physiol. Rev. 81, 971–998 (2001).
Moriyoshi, K. et al. Molecular cloning and characterization of the rat NMDA receptor. Nature 354, 31–37 (1991).
Meguro, H. et al. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature 357, 70–74 (1992).
Monyer, H. et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science 256, 1217–1221 (1992).
Kutsuwada, T. et al. Molecular diversity of the NMDA receptor channel. Nature 358, 36–41 (1992).
Ishii, T. et al. Molecular characterization of the family of the N-methyl-d-aspartate receptor subunits. J. Biol. Chem. 268, 2836–2843 (1993).
Ciabarra, A. M. et al. Cloning and characterization of χ-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J. Neurosci. 15, 6498–6508 (1995).
Sucher, N. J. et al. Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J. Neurosci. 15, 6509–6520 (1995).
Nishi, M., Hinds, H., Lu, H.-P., Kawata, M. & Hayashi, Y. Motoneuron-specific expression of NR3B, a novel NMDA-type glutamate receptor subunit that works in a dominant-negative manner. J. Neurosci. 21, RC185 (2001).
Johnson, J. W. & Ascher, P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature 325, 529–531 (1987).
Mothet, J. P. et al. d-serine is an endogenous ligand for the glycine site of the N-methyl-d-aspartate receptor. Proc. Natl Acad. Sci. USA 97, 4926–4931 (2000).
Nowak, L., Bregestovski, P., Ascher, P., Herbet, A. & Prochiantz, A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature 307, 462–465 (1984).
Mayer, M. L., Westbrook, G. L. & Guthrie, P. B. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309, 261–263 (1984).
Wenthold, R. J., Yokotani, N., Doi, K. & Wada, K. Immunochemical characterization of the non-NMDA glutamate receptor using subunit-specific antibodies. Evidence for a hetero-oligomeric structure in rat brain. J. Biol. Chem. 267, 501–507 (1992).
Brose, N. et al. Differential assembly of coexpressed glutamate receptor subunits in neurons of rat cerebral cortex. J. Biol. Chem. 269, 16780–16784 (1994).
Puchalski, R. B. et al. Selective RNA editing and subunit assembly of native glutamate receptors. Neuron 13, 131–147 (1994).
Seeburg, P. H. The TINS/TiPS lecture. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 16, 359–365 (1993).
Lomeli, H. et al. The rat δ-1 and δ-2 subunits extend the excitatory amino acid receptor family. FEBS Lett. 315, 318–322 (1993).
Kashiwabuchi, N. et al. Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR mutant mice. Cell 81, 245–252 (1995).
Schuster, C. M. et al. Molecular cloning of an invertebrate glutamate receptor subunit expressed in Drosophila muscle. Science 254, 112–114 (1991).
Maricq, A. V., Peckol, E., Driscoll, M. & Bargmann, C. I. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 378, 78–81 (1995).
Lam, H.-M. et al. Glutamate-receptor genes in plants. Nature 396, 125–126 (1998).
Chiu, J., DeSalle, R., Lam, H. M., Meisel, L. & Coruzzi, G. Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged. Mol. Biol. Evol. 16, 826–838 (1999).
Patneau, D. K. & Mayer, M. L. Structure–activity relationships for amino acid transmitter candidates acting at N-methyl-d-aspartate and quisqualate receptors. J. Neurosci. 10, 2385–2399 (1990).
Acknowledgements
This review is dedicated to the memory of Don C. Wiley, a great scientist, mentor and friend.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
1BL8: potassium channel (KcsA)
1EWK: mGluR subtype 1, complex with glutamate
1FTJ: GluR2 S1S2J, complex with glutamate
1FTK: GluR2 S1S2I, complex with kainate
1FTL: GluR2 S1S2J, complex with DNQX
1FTM: GluR2 S1S2J, complex with AMPA
1FW0: GluR2 S1S2J, complex with kainate
1IIW: GluR0 ligand-binding core, apo state
FURTHER INFORMATION
Families of Transport Proteins
Ion Channel Structure Research Group
Glossary
- ALLOSTERIC
-
A term originally used to describe enzymes that have two or more receptor sites, one of which (the active site) binds the principal substrate, whereas the other(s) bind(s) effector molecules that can influence its biological activity. More generally, it is used to describe the indirect coupling of distinct sites within a protein, mediated by conformational changes.
- P-LOOP SEQUENCE
-
A conserved structural motif found in many different ion channels that constitutes part of the channel pore.
- PDZ DOMAIN
-
A peptide-binding domain that is important for the organization of membrane proteins, particularly at cell–cell junctions, including synapses. They can bind to the carboxyl termini of proteins or can form dimers with other PDZ domains. PDZ domains are named after the proteins in which these sequence motifs were originally identified (PSD95, Discs large, zona occludens 1).
- STOPPED-FLOW FLUORESCENCE STUDIES
-
The properties of a fluorophore are dependent on its chemical environment, and can therefore be influenced by molecular interactions or conformational changes. In stopped-flow experiments, interacting molecules are rapidly mixed in a flow cell, and the temporal evolution of their fluorescence signals is monitored to determine the kinetics of the interaction.
- LURCHER MUTATION
-
A spontaneous gain-of-function mutation in the δ2 glutamate receptor gene of mice that causes neuronal cell death, leading to loss of motor control in heterozygotes and to early postnatal mortality in homozygotes.
- INFRARED SPECTROSCOPY
-
Many interatomic molecular vibrations show energy transitions that lead to the absorption of photons at characteristic frequencies in the infrared range. The energy transitions, and therefore the associated infrared absorption spectra, are highly sensitive to the stereochemical environment of the functional group(s) involved.
- ELECTRON PARAMAGNETIC RESONANCE SPECTROSCOPY
-
When an atom with an unpaired electron is placed in a magnetic field, the spin of the unpaired electron can align, either in the same direction as the field or in the opposite direction. Electron paramagnetic resonance (EPR) spectroscopy is used to measure the absorption of microwave radiation that accompanies the transition between these two states.
- EC50
-
The concentration of agonist that evokes a half-maximal response.
- INHIBITION CONSTANT
-
A measure of affinity, determined by the displacement of a labelled reporter ligand.
Rights and permissions
About this article
Cite this article
Madden, D. The structure and function of glutamate receptor ion channels. Nat Rev Neurosci 3, 91–101 (2002). https://doi.org/10.1038/nrn725
Issue Date:
DOI: https://doi.org/10.1038/nrn725
This article is cited by
-
Effects of chronic methamphetamine exposure on rewarding behavior and neurodegeneration markers in adult mice
Psychopharmacology (2023)
-
Protopanaxadiol ginsenoside Rd protects against NMDA receptor-mediated excitotoxicity by attenuating calcineurin-regulated DAPK1 activity
Scientific Reports (2020)
-
Genetic Mutation of GluN2B Protects Brain Cells Against Stroke Damages
Molecular Neurobiology (2018)
-
Incorporation of dihydroartemisinin into memantine through a propriate spacer to make hybrid with enhanced effects to protect PC12 cells from corticosterone-caused impairments
Chemical Research in Chinese Universities (2017)
-
Tissue Plasminogen Activator Neurotoxicity is Neutralized by Recombinant ADAMTS 13
Scientific Reports (2016)