Key Points
-
Multisubunit RNA polymerases (RNAPs) from the three domains of life are evolutionarily related, and this is reflected in the sequence and structure of their subunits, their interactions with transcription factors and their molecular mechanisms of action.
-
RNAP subunits contribute in distinct ways to enzyme function and facilitate assembly, catalysis, interaction with DNA and RNA, and regulation of RNAP activity.
-
The Rpo4–Rpo7 subunits of archaeal RNAP and the RPB4–RPB7 subunits of eukaryotic RNAPs constitute the stalk, which modulates the elongation and termination properties of these RNAPs but is not conserved in bacteria.
-
RNAP function is dependent on and regulated by exogenous transcription factors, some of which are universally conserved in evolution.
-
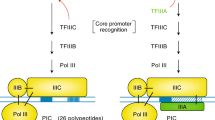
Transcription factors that enable transcription initiation in bacteria (σ-factors) and archaea and eukaryotes (TATA box-binding protein (TBP) and TFIIB) are not homologous.
-
Transcript cleavage factors in bacteria (GreB) and archaea and eukaryotes (TFIIS) are not homologous.
-
Non-homologous transcription factors can adapt a similar structure and can therefore interact with and regulate their cognate RNAPs using closely related molecular mechanisms.
-
NusG is the only RNAP-associated transcription factor that is universally conserved in evolution (NusG in bacteria, Spt5 in archaea and SPT5 in eukaryotes), indicating that regulation of transcription during elongation predated regulation of transcription initiation.
Abstract
RNA polymerases (RNAPs) carry out transcription in all living organisms. All multisubunit RNAPs are derived from a common ancestor, a fact that becomes apparent from their amino acid sequence, subunit composition, structure, function and molecular mechanisms. Despite the similarity of these complexes, the organisms that depend on them are extremely diverse, ranging from microorganisms to humans. Recent findings about the molecular and functional architecture of RNAPs has given us intriguing insights into their evolution and how their activities are harnessed by homologous and analogous basal factors during the transcription cycle. We provide an overview of the evolutionary conservation of and differences between the multisubunit polymerases in the three domains of life, and introduce the 'elongation first' hypothesis for the evolution of transcriptional regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Werner, F. Structural evolution of multisubunit RNA polymerases. Trends Microbiol. 16, 247–250, (2008).
Werner, F. Structure and function of archaeal RNA polymerases. Mol. Microbiol. 65, 1395–1404 (2007).
Cramer, P. et al. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys. 37, 337–352 (2008).
Cheetham, G. M. & Steitz, T. A. Insights into transcription: structure and function of single-subunit DNA-dependent RNA polymerases. Curr. Opin. Struct. Biol. 10, 117–123 (2000).
Steitz, T. A. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr. Opin. Struct. Biol. 19, 683–690 (2009).
Cramer, P., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: RNA polymerase II at 2.8 Ångstrom resolution. Science 292, 1863–1876 (2001).
Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292, 1876–1882 (2001).
Hirata, A., Klein, B. J. & Murakami, K. S. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451, 851–854 (2008).
Korkhin, Y. et al. Evolution of complex RNA polymerases: the complete archaeal RNA polymerase structure. PLoS Biol. 7, e102 (2009).
Zhang, G. et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98, 811–824 (1999).
Hirtreiter, A., Grohmann, D. & Werner, F. Molecular mechanisms of RNA polymerase — the F/E (RPB4/7) complex is required for high processivity in vitro. Nucleic Acids Res. 38, 585–596 (2010).
Batada, N. N., Westover, K. D., Bushnell, D. A., Levitt, M. & Kornberg, R. D. Diffusion of nucleoside triphosphates and role of the entry site to the RNA polymerase II active center. Proc. Natl Acad. Sci. USA 101, 17361–17364 (2004).
Iyer, L. M., Koonin, E. V. & Aravind, L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 3, 1 (2003).
Grohmann, D., Werner, F. Cycling through transcription with the RNA polymerase F/E (RPB4/7) complex: structure, function and evolution of archaeal RNA polymerase. Res. Microbiol. 21 Sep 2010 (doi: 10.1016/j.resmic.2010.09.002).
Wassarman, K. M. & Saecker, R. M. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 314, 1601–1603 (2006).
Lehmann, E., Brueckner, F. & Cramer, P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature 450, 445–449 (2007).
Severinov, K. et al. Structural modules of the large subunits of RNA polymerase. Introducing archaebacterial and chloroplast split sites in the β and β′ subunits of Escherichia coli RNA polymerase. J. Biol. Chem. 271, 27969–27974 (1996).
Zakharova, N. et al. Fused and overlapping rpoB and rpoC genes in helicobacters, campylobacters, and related bacteria. J. Bacteriol. 181, 3857–3859 (1999).
Severinov, K., Mooney, R., Darst, S. A. & Landick, R. Tethering of the large subunits of Escherichia coli RNA polymerase. J. Biol. Chem. 272, 24137–24140 (1997).
Sweetser, D., Nonet, M. & Young, R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc. Natl Acad. Sci. USA 84, 1192–1196 (1987).
Lane, W. J. & Darst, S. A. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 395, 671–685 (2010).
Lane, W. J. & Darst, S. A. Molecular evolution of multisubunit RNA polymerases: structural analysis. J. Mol. Biol. 395, 686–704 (2010).
Ruprich-Robert, G. & Thuriaux, P. Non-canonical DNA transcription enzymes and the conservation of two-barrel RNA polymerases. Nucleic Acids Res. 38, 4559–4569 (2010).
Makeyev, E. V. & Bamford, D. H. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell 10, 1417–1427 (2002).
Salgado, P. S. et al. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 4, e434 (2006).
Orlicky, S. M., Tran, P. T., Sayre, M. H. & Edwards, A. M. Dissociable Rpb4-Rpb7 subassembly of RNA polymerase II binds to single-strand nucleic acid and mediates a post-recruitment step in transcription initiation. J. Biol. Chem. 276, 10097–10102 (2001).
Grohmann, D., Hirtreiter, A. & Werner, F. RNAP subunits F/E (RPB4/7) are stably associated with archaeal RNA polymerase: using fluorescence anisotropy to monitor RNAP assembly in vitro. Biochem. J. 421, 339–343 (2009).
Grohmann, D., Werner, F. Hold on! RNAP interactions with the nascent RNA modulate transcription elongation and termination. RNA Biol. 7, 310–315 (2010).
Naji, S., Grunberg, S. & Thomm, M. The RPB7 orthologue E′ is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J. Biol. Chem. 282, 11047–11057 (2007).
Ouhammouch, M., Werner, F., Weinzierl, R. O. & Geiduschek, E. P. A fully recombinant system for activator-dependent archaeal transcription. J. Biol. Chem. 279, 51719–51721 (2004).
Werner, F. & Weinzierl, R. O. Direct modulation of RNA polymerase core functions by basal transcription factors. Mol. Cell Biol. 25, 8344–8355 (2005).
Werner, F. & Weinzierl, R. O. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10, 635–646 (2002).
Werner, F., Eloranta, J. J. & Weinzierl, R. O. Archaeal RNA polymerase subunits F and P are bona fide homologs of eukaryotic RPB4 and RPB12. Nucleic Acids Res. 28, 4299–4305 (2000).
Kostrewa, D. et al. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009).
Reich, C. et al. The archaeal RNA polymerase subunit P and the eukaryotic polymerase subunit Rpb12 are interchangeable in vivo and in vitro. Mol. Microbiol. 71, 989–1002 (2009).
Grunberg, S., Reich, C., Zeller, M. E., Bartlett, M. S. & Thomm, M. Rearrangement of the RNA polymerase subunit H and the lower jaw in archaeal elongation complexes. Nucleic Acids Res. 38, 1950–1963 (2010).
Bartlett, M. S., Thomm, M. & Geiduschek, E. P. Topography of the euryarchaeal transcription initiation complex. J. Biol. Chem. 279, 5894–5903 (2004).
Kang, X. et al. Structural, biochemical, and dynamic characterizations of the hRPB8 subunit of human RNA polymerases. J. Biol. Chem. 281, 18216–18226 (2006).
Komissarova, N. & Kashlev, M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl Acad. Sci. USA 95, 14699–14704 (1998).
Briand, J. F. et al. Partners of Rpb8p, a small subunit shared by yeast RNA polymerases I, II and III. Mol. Cell Biol. 21, 6056–6065 (2001).
Cox, C. J., Foster, P. G., Hirt, R. P., Harris, S. R. & Embley, T. M. The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA 105, 20356–20361 (2008).
Walmacq, C. et al. Rpb9 subunit controls transcription fidelity by delaying NTP sequestration in RNA polymerase II. J. Biol. Chem. 284, 19601–19612 (2009).
Ziegler, L. M., Khaperskyy, D. A., Ammerman, M. L. & Ponticelli, A. S. Yeast RNA polymerase II lacking the Rpb9 subunit is impaired for interaction with transcription factor IIF. J. Biol. Chem. 278, 48950–48956 (2003).
Campbell, E. A., Westblade, L. F. & Darst, S. A. Regulation of bacterial RNA polymerase σ factor activity: a structural perspective. Curr. Opin. Microbiol. 11, 121–127 (2008).
Murakami, K. S. & Darst, S. A. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13, 31–39 (2003).
Gruber, T. M. & Gross, C. A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466 (2003).
MacLellan, S. R., Guariglia-Oropeza, V., Gaballa, A. & Helmann, J. D. A two-subunit bacterial σ-factor activates transcription in Bacillus subtilis. Proc. Natl Acad. Sci. USA 106, 21323–21328 (2009).
MacLellan, S. R., Wecke, T. & Helmann, J. D. A previously unidentified σ factor and two accessory proteins regulate oxalate decarboxylase expression in Bacillus subtilis. Mol. Microbiol. 69, 954–967 (2008).
Wigneshweraraj, S. et al. Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol. Microbiol. 68, 538–546 (2008).
Kim, T. K., Ebright, R. H. & Reinberg, D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288, 1418–1422 (2000).
Schwartz, E. C. et al. A full-length group 1 bacterial sigma factor adopts a compact structure incompatible with DNA binding. Chem. Biol. 15, 1091–1103 (2008).
Patikoglou, G. A. et al. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13, 3217–3230 (1999).
Campbell, E. A. et al. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell 9, 527–539 (2002).
Chen, H. T. & Hahn, S. Binding of TFIIB to RNA polymerase II: mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell 12, 437–447 (2003).
Chen, H. T. & Hahn, S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell 119, 169–180 (2004).
Sevostyanova, A., Svetlov, V., Vassylyev, D. G. & Artsimovitch, I. The elongation factor RfaH and the initiation factor σ bind to the same site on the transcription elongation complex. Proc. Natl Acad. Sci. USA 105, 865–870 (2008).
Renfrow, M. B. et al. Transcription factor B contacts promoter DNA near the transcription start site of the archaeal transcription initiation complex. J. Biol. Chem. 279, 2825–2831 (2004).
Bushnell, D. A., Westover, K. D., Davis, R. E. & Kornberg, R. D. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science 303, 983–988 (2004).
Liu, X., Bushnell, D. A., Wang, D., Calero, G. & Kornberg, R. D. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 327, 206–209 (2010).
Kulbachinskiy, A. & Mustaev, A. Region 3.2 of the σ subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 281, 18273–18276 (2006).
Kapanidis, A. N. et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314, 1144–1147 (2006).
Goldman, S. R., Ebright, R. H. & Nickels, B. E. Direct detection of abortive RNA transcripts in vivo. Science 324, 927–928 (2009).
Murakami, K. S., Masuda, S., Campbell, E. A., Muzzin, O. & Darst, S. A. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296, 1285–1290 (2002).
Holstege, F. C., van der Vliet, P. C. & Timmers, H. T. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15, 1666–1677 (1996).
Ohkuma, Y. et al. Structural motifs and potential σ homologies in the large subunit of human general transcription factor TFIIE. Nature 354, 398–401 (1991).
Sumimoto, H. et al. Conserved sequence motifs in the small subunit of human general transcription factor TFIIE. Nature 354, 401–404 (1991).
Garrett, K. P. et al. The carboxyl terminus of RAP30 is similar in sequence to region 4 of bacterial sigma factors and is required for function. J. Biol. Chem. 267, 23942–23949 (1992).
Flores, O. et al. The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc. Natl Acad. Sci. USA 88, 9999–10003 (1991).
Yan, Q., Moreland, R. J., Conaway, J. W. & Conaway, R. C. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J. Biol. Chem. 274, 35668–35675 (1999).
Eichner, J., Chen, H. T., Warfield, L. & Hahn, S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 29, 706–716 (2010).
Geiger, S. R. et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell 39, 583–594 (2010).
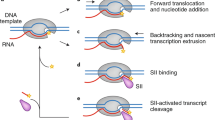
Selth, L. A., Sigurdsson, S. & Svejstrup, J. Q. Transcript elongation by RNA polymerase II. Annu. Rev. Biochem. 79, 271–293 (2010).
Vassylyev, D. G. Elongation by RNA polymerase: a race through roadblocks. Curr. Opin. Struct. Biol. 19, 691–700 (2009).
Landick, R. The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 34, 1062–1066 (2006).
Brueckner, F., Ortiz, J. & Cramer, P. A movie of the RNA polymerase nucleotide addition cycle. Curr. Opin. Struct. Biol. 19, 294–299 (2009).
Brueckner, F. et al. Structure–function studies of the RNA polymerase II elongation complex. Acta Crystallogr. D Biol. Crystallogr. 65, 112–120 (2009).
Ha, K. S., Toulokhonov, I., Vassylyev, D. G. & Landick, R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J. Mol. Biol. 401, 708–725 (2010).
Shibata, R. et al. Crystal structure and RNA-binding analysis of the archaeal transcription factor NusA. Biochem. Biophys. Res. Commun. 355, 122–128 (2007).
Beuth, B., Pennell, S., Arnvig, K. B., Martin, S. R. & Taylor, I. A. Structure of a Mycobacterium tuberculosis NusA–RNA complex. EMBO J. 24, 3576–3587 (2005).
Runner, V. M., Podolny, V. & Buratowski, S. The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3 processing factors. Mol. Cell Biol. 28, 1883–1891 (2008).
Armache, K. J., Mitterweger, S., Meinhart, A. & Cramer, P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 280, 7131–7134 (2005).
Deighan, P. & Hochschild, A. Conformational toggle triggers a modulator of RNA polymerase activity. Trends Biochem. Sci. 31, 424–426 (2006).
Sigurdsson, S., Dirac-Svejstrup, A. B. & Svejstrup, J. Q. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol. Cell 38, 202–210 (2010).
Hausner, W., Lange, U. & Musfeldt, M. Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J. Biol. Chem. 275, 12393–12399 (2000).
Laptenko, O., Lee, J., Lomakin, I. & Borukhov, S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 22, 6322–6334 (2003).
Kettenberger, H., Armache, K. J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965 (2004).
Jeon, C., Yoon, H. & Agarwal, K. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc. Natl Acad. Sci. USA 91, 9106–9110 (1994).
Borukhov, S., Lee, J. & Laptenko, O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol. Microbiol. 55, 1315–1324 (2005).
Opalka, N. et al. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114, 335–345 (2003).
Condon, C., Squires, C. & Squires, C. L. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59, 623–645 (1995).
Arnvig, K. B. et al. Evolutionary comparison of ribosomal operon antitermination function. J. Bacteriol. 190, 7251–7257 (2008).
Greenblatt, J., Nodwell, J. R. & Mason, S. W. Transcriptional antitermination. Nature 364, 401–406 (1993).
Dodd, I. B., Shearwin, K. E. & Egan, J. B. Revisited gene regulation in bacteriophage l. Curr. Opin. Genet. Dev. 15, 145–152 (2005).
Hirtreiter, A. et al. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled coil motif. Nucleic Acids Res. 38, 4040–4051 (2010).
Mooney, R. A., Schweimer, K., Roesch, P., Gottesman, M. & Landick, R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2, 341–358 (2009).
Zhou, K., Kuo, W. H., Fillingham, J. & Greenblatt, J. F. Control of transcriptional elongation and cotranscriptional histone modification by the yeast BUR kinase substrate Spt5. Proc. Natl Acad. Sci. USA 106, 6956–6951 (2009).
Nickels, B. E., Mukhopadhyay, J., Garrity, S. J., Ebright, R. H. & Hochschild, A. The s70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nature Struct. Mol. Biol. 11, 544–550 (2004).
Artsimovitch, I. Post-initiation control by the initiation factor sigma. Mol. Microbiol. 68, 1–3 (2008).
Missra, A. & Gilmour, D. S. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc. Natl Acad. Sci. USA 107, 11301–11306 (2010).
Cheng, B. & Price, D. H. Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay. Nucleic Acids Res. 36, e135 (2008).
Svetlov, V., Belogurov, G. A., Shabrova, E., Vassylyev, D. G. & Artsimovitch, I. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 35, 5694–5705 (2007).
Belogurov, G. A. et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell 26, 117–129 (2007).
Tan, L., Wiesler, S., Trzaska, D., Carney, H. C. & Weinzierl, R. O. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J. Biol. 7, 40 (2008).
Peters, J. P. 3rd & Maher, L. J. DNA curvature and flexibility in vitro and in vivo. Q. Rev. Biophys. 43, 23–63 (2010).
Herbert, K. M. et al. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J. Mol. Biol. 399, 17–30 (2010).
Proshkin, S., Rahmouni, A. R., Mironov, A. & Nudler, E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328, 504–508 (2010).
Burmann, B. M. et al. A NusE:NusG complex links transcription and translation. Science 328, 501–504 (2010).
Vassylyev, D. G., Vassylyeva, M. N., Perederina, A., Tahirov, T. H. & Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 (2007).
Andrecka, J. et al. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic Acids Res. 37, 5803–5809 (2009).
Greive, S. J. & von Hippel, P. H. Thinking quantitatively about transcriptional regulation. Nature Rev. Mol. Cell Biol. 6, 221–232 (2005).
Wang, Z., Gerstein, M. & Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Rev. Genet. 10, 57–63 (2009).
Mooney, R. A. et al. Regulator trafficking on bacterial transcription units in vivo. Mol. Cell 33, 97–108 (2009).
Sutherland, H. & Bickmore, W. A. Transcription factories: gene expression in unions? Nature Rev. Genet. 10, 457–466 (2009).
Epshtein, V., Toulme, F., Rahmouni, A. R., Borukhov, S. & Nudler, E. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 22, 4719–4727 (2003).
Saeki, H. & Svejstrup, J. Q. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol. Cell 35, 191–205 (2009).
Werner, M., Thuriaux, P. & Soutourina, J. Structure–function analysis of RNA polymerases I and III. Curr. Opin. Struct. Biol. 19, 740–745 (2009).
Spitalny, P. & Thomm, M. A polymerase III-like reinitiation mechanism is operating in regulation of histone expression in archaea. Mol. Microbiol. 67, 958–970 (2008).
Landick, R. Functional divergence in the growing family of RNA polymerases. Structure 17, 323–325 (2009).
Vanhamme, L. Trypanosome RNA polymerases and transcription factors: sensible trypanocidal drug targets? Curr. Drug Targets. 9, 979–996 (2008).
Grunberg, S., Bartlett, M. S., Naji, S. & Thomm, M. Transcription factor E is a part of transcription elongation complexes. J. Biol. Chem. 282, 35482–35490 (2007).
Acknowledgements
We thank S. Gribaldo from the Institute Pasteur, Paris, France, for stimulating discussions on evolution and the inspiration for figure 8. We also thank K. S. Murakami for sharing unpublished results.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Glossary
- Last universal common ancestor
-
The last common ancestor of all three contemporary domains of life.
- Transcription factor
-
A protein that transiently interacts with RNA polymerase, modulates its protein-, DNA- and RNA-binding or catalytic properties, and thereby regulates transcription.
- Convergent evolution
-
The acquisition of the same trait in unrelated lineages.
- Homologous
-
Pertaining to genes: derived from the same ancestor.
- RNAP clamp
-
A flexible RNA polymerase (RNAP) domain that is predicted to move over the DNA-binding channel during open-complex formation.
- Bridge and trigger helices
-
Flexible RNA polymerase motifs that unfold and refold repeatedly during nucleotide addition and translocation.
- Double-psi β-barrel motif
-
A structural module in the two largest RNA polymerase subunits that reveals the common origin of these subunits and constitutes the active site.
- Ribozyme
-
An enzyme made exclusively of RNA.
- RNAi RNAP
-
An RNA polymerase (RNAP) involved in RNA interference (RNAi); it produces double-stranded RNA from aberrant single-stranded RNA templates to trigger RNAi.
- Open-complex formation
-
The structural transition of RNA polymerase concomitant with the melting of DNA strands during initiation.
- TATA box-binding protein
-
One of the two minimal transcription factors required for initiation by archaeal RNA polymerase (RNAP) and eukaryotic RNAPII. It binds to a sequence recognition motif in promoters that is called the TATA element or TATA box.
- TFIIB
-
The second of the two minimal transcription factors required for initiation by archaeal RNA polymerase (RNAP) and eukaryotic RNAPII.
- σ-factor
-
A bacterial transcription initiation factor that binds specific sequences in the promoter. Each σ-factor regulates the transcription of a specific set of genes.
- −10 and −35 elements
-
Key sequence motifs of bacterial promoters that are recognized by the transcription factor σ70. Promoter strength is in part regulated by the strength of the interaction between σ70 and these elements.
- Zn-ribbon domain
-
A domain found in many transcription factors, including TFIIB, TFIIS and TFIIE in eukaryotes (and TFB, TFS and TFE, respectively, in archaea). The aminoterminal Zn-ribbon domain of TFIIB and TFB interacts with the RNA polymerase (RNAP) dock domain and is important for efficient RNAP recruitment.
- Paralogous
-
Pertaining to genes: separated by a gene duplication event.
Rights and permissions
About this article
Cite this article
Werner, F., Grohmann, D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 9, 85–98 (2011). https://doi.org/10.1038/nrmicro2507
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2507
This article is cited by
-
Tail-tape-fused virion and non-virion RNA polymerases of a thermophilic virus with an extremely long tail
Nature Communications (2024)
-
A functional promoter from the archaeon Halobacterium salinarum is also transcriptionally active in E. coli
BMC Microbiology (2022)
-
Promoter-proximal elongation regulates transcription in archaea
Nature Communications (2021)
-
Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II
Nature Chemical Biology (2021)
-
Structure and function of virion RNA polymerase of a crAss-like phage
Nature (2021)