Key Points
-
This Review focuses on the mechanisms and clinical importance of the bacterial–fungal interactions that occur on or in the human body.
-
Bacteria and fungi can interact in several ways, including physical interactions by direct cell–cell contact, chemical interaction through the secretion of small molecules that are often involved in quorum sensing, environmental modifications such as pH changes, use of metabolic by-products and alterations in host responses.
-
A range of mammalian and non-mammalian models of infection are now available for the study of mixed bacterial–fungal infections.
-
Several Gram-negative pathogens are capable of killing Candida albicans and inhibiting filament formation, including Pseudomonas aeruginosa, Acinetobacter baumannii, Burkholderia cepacia and Salmonella enterica subsp. enterica serovar Typhimurium. This is predominantly mediated through the secretion of small molecules, such as quorum-sensing molecules and other known secretory virulence factors (namely, phospholipase C and phenazines for P. aeruginosa).
-
Oral streptococci have adapted to adhere to C. albicans in the human mouth, and this seems to be mediated through polysaccharide receptors on the bacterial surface. Such co-aggregation is important in the pathogenesis of many oral diseases.
-
C. albicans mounts a defence against these bacterial predators through the secretion of its quorum-sensing molecule farnesol. This molecule can affect bacterial production of virulence factors, viability and susceptibility to antibacterials.
-
Limited study has been dedicated to understanding the host responses to polymicrobial infections. Recent work in mice suggests that immune responses to a bacterial–fungal infection may be directed preferentially towards a bacterial-type response mediated by T helper 1 cells.
Abstract
Whether it is in the setting of disease or in a healthy state, the human body contains a diverse range of microorganisms, including bacteria and fungi. The interactions between these taxonomically diverse microorganisms are highly dynamic and dependent on a multitude of microorganism and host factors. Human disease can develop from an imbalance between commensal bacteria and fungi or from invasion of particular host niches by opportunistic bacterial and fungal pathogens. This Review describes the clinical and molecular characteristics of bacterial–fungal interactions that are relevant to human disease.
Similar content being viewed by others
Main
Despite the abundance of bacterial–fungal interactions in nature and the clinical environment, very little is known about the molecular mechanisms underlying these interactions and their importance to human health. Microorganisms have evolved complex mechanisms to promote their survival, defending themselves not only against adverse environmental or nutritional conditions but also against competing organisms. Unravelling the mechanisms that microorganisms use in a competitive, polymicrobial environment would not only deepen our understanding of microbial pathogenesis but may also provide important insights into novel pathways that are amenable for the development of new antimicrobial drugs. History has demonstrated the power of understanding such interactions, with the identification of penicillin being the consequence of a bacterial–fungal interaction on a contaminated agar plate1.
This Review will describe the mechanisms and medical importance of bacterial–fungal interactions that occur in or on the human body. Owing to the extensive distribution of the fungal species Candida albicans on human skin and mucosal surfaces, this is the fungus that is most frequently implicated in mixed bacterial–fungal infections and will therefore be the main focus of this Review. In addition, the interactions of other fungi such as Cryptococcus neoformans with bacteria will be explored.
Sites of bacterial–fungal interactions
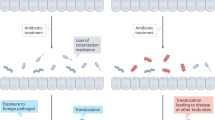
In the absence of disease, bacteria and fungi are most commonly found on cutaneous and mucosal surfaces such as the skin, the oral cavity, the gastrointestinal tract and the lower female reproductive tract. Localized insults, such as burn injury to the skin, poor dental hygiene, a surgical procedure, or oral or gastrointestinal mucositis as a consequence of chemotherapy, can lead to disease at these sites, and such diseases are often polymicrobial in nature2,3,4 (Fig. 1). Breaches in tissue barriers can also lead to the expansion of these organisms into normally sterile sites such as the bloodstream5,6,7. Similarly, systemic insults to human microbial ecology such as antimicrobial therapy or deficiencies in host immunity can lead to an imbalance in the normal microbial flora and allow a normally benign, commensal organism to become pathogenic, as is the case in vaginal candidiasis, which can occur after the use of systemic antibacterials. Furthermore, colonization of the respiratory tract by bacteria and fungi is especially frequent in patients with chronic lung diseases8,9 and in mechanically ventilated patients in intensive care units10,11 (Fig. 1), where mixed bacterial–fungal biofilms are commonly found12,13. In-dwelling medical devices that breach the skin can be similarly affected by polymicrobial biofilms7,14.
Mechanisms of interaction
Bacteria and fungi directly and indirectly influence each other in several ways (Fig. 2). Bacterial factors can influence fungal growth or physiology, and, conversely, fungal factors have been shown to control bacterial behaviour and survival. The virulence of bacteria15 or fungi16 can also be influenced by a polymicrobial encounter. Strong evidence indicates that secreted molecules mediate many types of interactions between bacteria and fungi. Interestingly, such extracellular signalling molecules often mediate quorum sensing in single-species communities, suggesting that the effects of one organism on another may be dependent on the population density15,17,18,19,20,21,22. In addition, bacterial toxins such as pseudomonal phenazines have been shown to have antifungal properties23,24,25,26.
Bacteria and fungi influence each other in many ways and can impact each other's survival or virulence15,16. The consequences of these interactions are similarly varied. Such associations can be either beneficial, as in the case of probiotic bacteria that decrease fungal colonization, or detrimental, as can be the case with mixed-species biofilms16. a | Physical interactions include bacterial attachment to the fungal surface or co-aggregation with fungal cells16,74,93, the formation of bacterial biofilms on the surface of fungal hyphae24 and the formation of mixed-species biofilms on an abiotic or host surface77. b | Chemical exchanges. For example, diverse bacteria produce small molecules that affect the morphology of Candida albicans, thereby altering the ability of the fungus to form biofilms or to invade tissues17,19,22. c | Use of metabolic by-products. d | Changes in the environment. e | Alteration of the host immune response. QS, quoroum-sensing; TH, T helper cell.
Diverse physical interactions between bacteria and fungi have also been described, ranging from bacterial cell contact and aggregation with fungal hyphae or yeast cells16,27 to organized bacterial biofilms on the surface of fungal hyphae24 (Fig. 2a). Such cellular interactions have been associated with reduced fungal viability, which may be mediated by bacterial secretion of antifungal molecules into the local environment, by the transfer of toxins directly into the fungal cell through secretion systems or by nutrient depletion. Another mechanism of bacterial–fungal interaction is environmental modification, such as a change in pH28, which can influence the formation of hyphae in C. albicans29. In addition to antagonistic interactions, mutually beneficial interactions in mixed-biofilm environments are also possible, whereby the different species may provide protection for each other against an attacking immune response or antimicrobial agent.
Clinical importance
Although data on the clinical relevance of bacterial–fungal interactions are limited, several studies have described the association of bacteria and Candida spp. in a range of clinical specimens4,12,30. It is not yet clear whether factors such as systemic antibacterial therapy, host immune status or exposure to hospital-acquired pathogens simply predispose a patient to colonization by both bacteria and fungi. However, mixed-species infections can have consequences that differ from those associated with single-species infections. A study of ventilator-associated pneumonia (VAP) caused by Pseudomonas aeruginosa suggested that colonization of the respiratory tract with Candida spp. may increase the risk of pseudomonal VAP11. In support of these observations, other studies have shown that, for individuals with tracheobronchial colonization by Candida spp., those who were treated with antifungal drugs had a lower risk of pseudomonal VAP than those who were not31. Data from animal models provide additional support for these findings, as described below.
Retrospective human studies suggest that mortality from bloodstream infections that are caused by bacteria or Candida spp. ranges from 10–40%32. However, very few studies have performed a comparison of single-species and mixed-species infections. One such study identified a poorer survival rate for a mixed bacteria–Candida spp. bloodstream infection than for an infection with Candida spp. alone5. Analyses of the implications of mixed-species infections are limited, as prospective, randomized human trials are rarely possible and observational studies are confounded by the fact that patients with polymicrobial infections may have other risk factors that correlate with a poor clinical outcome, such as greater severity of illness or inadequate therapy against either or both infecting organisms. Furthermore, describing the molecular mechanisms by which any changes in virulence occur in a polymicrobial infection is difficult when studying human disease.
Virulence in animal model systems
An understanding of the pathogenic consequences of bacterial–fungal interactions has come predominately from research using mammalian models of infection (Box 1). Assessing the changes in virulence in a bacterial–fungal infection is complex and depends on several factors: the temporal association of the microbial encounter (whether the bacterial and fungal infections develop simultaneously, or the bacteria develop a quorum before the fungi or vice versa); the site of infection; the size of the inoculum; the organism type; the host; and the outcome measurement of interest. Common outcome measurements for virulence assessment in mammals include the change in the microbial burden at the primary site of infection, the rate of dissemination of the infection from the primary site, the microbial burden at secondary sites, tissue histopathology and host survival over time. Except for host survival, the methods used to study single-species microbial infections can be applied for all of these measurements. Survival-curve analysis and interpretation is more challenging when studying a polymicrobial infection with its single-species comparators and is described in detail in Box 2.
Enhanced fungal virulence. One of the earliest studies assessing the pathogenic consequences of a mixed bacterial–fungal infection assessed the interaction between C. albicans and Escherichia coli in mice33. Interestingly, attenuation in host killing by C. albicans was observed if a sublethal dose of E. coli was administered intravenously or intraperitoneally before a lethal dose of C. albicans. This mirrored in vitro data, which showed that E. coli was able to reduce the viability of C. albicans over time33. However, the mechanism behind these observations was not described. Conversely, if the E. coli was given after inoculation with C. albicans, enhanced killing was observed (for the most part), which was thought to be mediated by E. coliendotoxin33. Several other investigators have subsequently had similar results, whereby simultaneous infection with C. albicans and E. coli caused enhanced killing compared with killing by either organism alone34,35,36,37, and endotoxin seemed to be important for this enhanced virulence34,35. These findings have clinical importance, as E. coli and C. albicans are commensals of the human gastrointestinal tract and are often found in intra-abdominal and hospital-acquired bloodstream infections. These data also highlight the potential importance of adequate antibacterial therapy in the setting of invasive candidiasis.
A further example of the increased virulence of C. albicans in the presence of bacteria is shown with a pseudomonal burn wound infection model38. Life-threatening candidaemia in human burn victims is often preceded by bacterial infection, especially by P. aeruginosa. When burn wounds on mice were pre-infected with a sublethal inoculum of P. aeruginosa and then exposed to a sublethal inoculum of C. albicans, the mice had a mortality rate of 60%38. By contrast, burned mice that were infected with the same inocula individually had a mortality rate of < 10%. Interestingly, microbial burden studies of the burn wound and peripheral organs in co-infected mice showed that the deaths seemed to be due to C. albicans. Furthermore, the authors found that the pseudomonal proteolytic enzyme, elastase (LasB; also known as pseudolysin), was responsible for the increased C. albicans virulence, but the details of this mechanism were unclear. In a similar study using a rat pneumonia model, investigators found that rats given a subclinical dose of P. aeruginosa developed pneumonia only in the presence of viable C. albicans39. These data support the clinical studies of pseudomonal VAP that were described above11,31.
A more recent study may shed some light on the possible mechanism behind the enhanced virulence of C. albicans in the setting of a bacterial infection. It was found that bacterial peptidoglycan-like molecules found in human serum, known as muramyl dipeptides, act as potent inducers for hyphal development in C. albicans40. The formation of hyphae is a crucial virulence determinant for C. albicans in mammalian infection41. The muramyl dipeptides are thought to originate from commensal bacteria, and it is therefore plausible that systemic bacterial infection leads to a large quantity of these hyphae-promoting molecules that subsequently enhance the virulence of C. albicans.
Enhanced bacterial virulence. There is also evidence for the enhancement of bacterial virulence by C. albicans, illustrating the dynamic nature of polymicrobial infections. This is well described in studies assessing the virulence of mixed C. albicans and Staphylococcus aureus infection in mice42,43. When S. aureus alone was inoculated intraperitoneally, no organisms could be identified in peripheral sites such as the blood, pancreas or spleen and the mice survived. However, when the same dose of S. aureus was administered with a sublethal dose of C. albicans or with heat-inactivated C. albicans, S. aureus was recovered 48 hours later in all samples of blood and abdominal organs and most of the mice died43. There was no difference observed in the fungal burden of organs for infections with C. albicans alone or together with S. aureus. Interestingly, histopathology of the peritoneal cavity indicated that S. aureus was always found at sites of fungal growth, even when the two pathogens were injected at different sites. Furthermore, in the setting of mixed infection, peripheral organs developed high staphylococcal burdens but negligible amounts of C. albicans43. These data suggest that C. albicans provides protection for S. aureus in the peritoneal cavity and enhances its virulence by allowing the bacteria to disseminate to peripheral tissues (a process that would not occur with S. aureus infection alone). However, the observed effects may be dependent on the bacterial strain44. Similar findings were observed by the same investigators for the Gram-negative bacterium Serratia marcescens and for another Gram-positive species, Enterococcus faecalis 42, which are both important human pathogens causing hospital-acquired infections that often originate from an intra-abdominal source, as found in the murine model. Given the importance of S. aureus as a human pathogen, its ability to cause life-threatening disseminated infections and its common co-habitation with C. albicans, these findings are of great importance to our understanding of S. aureus pathogenesis in humans. Furthermore, these data also suggest a potential indirect benefit of antifungal treatment in the setting of complex intra-abdominal bacterial–fungal infections: the reduction of bacterial virulence.
Host responses to mixed infections
Host immune responses to polymicrobial infections have so far been underappreciated. Cross-kingdom microbial interactions such as mixed bacterial–fungal infections challenge the immune system in diverse ways compared with infection with either organism alone. A recent study illustrates the clinical importance of host responses to mixed bacterial–fungal infections45. Using a mouse model, the immune responses to direct lung exposure to pseudomonal and/or C. albicans cellular lysates were assessed. As had been shown previously, exposure to the fungal antigens led to substantial airway inflammation characterized by eosinophilic infiltration, mucus production and an increase in levels of T helper 2 (TH2) cell cytokines (namely, interleukin-4 (IL-4), IL-5 and IL-13). Exposure to pseudomonal lysates also led to airway inflammation, but this was characterized by neutrophilic infiltration, no mucus production and an increase in the production of interferon-γ (IFNγ), which is a TH1 cell CD4+ T cell cytokine45. Interestingly, when mice were exposed to both fungal and bacterial lysates, similar degrees of airway inflammation were observed, but the immune response was neutrophilic rather than eosinophilic and TH1 cell rather than TH2 cell cytokines were produced45. Furthermore, on subsequent exposure to C. albicans lysates alone, the immune response had diverted and was characterized by neutrophils and TH1 cell cytokines. These data demonstrate that, at least in mice, the type of immune response and inflammation that is mounted to a mixed antigen load is dictated by the presence or absence of bacterial antigens. Whether the same results would be obtained using live microorganisms and involving other organ sites is currently unknown. The implications of these findings are broad and lend support to the 'hygiene hypothesis', whereby an increase in atopic disease in the developed world, mediated by an increase in TH2 cell responses, is related to a more hygienic environment in which there is less bacterial stimulation46,47,48. Bacterial exposure may reduce such responses through its activation of TH1 cells (Fig. 2e).
An altered host response to one pathogen may also promote the success of another39. In a rat lung model of C. albicans colonization and subsequent pseudomonal exposure, an increase in the rate of pseudomonal pneumonia was observed. Interestingly, production of reactive oxygen species by alveolar macrophages was notably inhibited by C. albicans39, suggesting that C. albicans may suppress local host immune responses to allow subclinical inocula of P. aeruginosa to cause disease.
Specific bacterial–fungal interactions
Interactions between Pseudomonas spp. and Candida spp. Many studies have described mixed infections with both P. aeruginosa and C. albicans in cases ranging from contaminated catheters to chronic lung infections4,8,49,50, and clinical observations suggest that the bacterial and fungal populations influence each other31,51,52. In vitro studies show the wide spectrum of interactions between P. aeruginosa and C. albicans (Fig. 3) and emphasize the importance of environmental conditions in determining the outcome of an interaction. For example, in liquid media, P. aeruginosa adheres to and forms biofilms on C. albicans filaments but not on yeast cells24 (Fig. 3a). This biofilm formation leads to the death of the fungal cell, brought about by the action of two pseudomonal virulence factors: a secreted haemolytic phospholipase C that degrades phosphatidylcholine (an abundant phospholipid in eukaryotes), and redox-active phenazines, which generate highly toxic reactive oxygen species23,24. Although biofilm formation is required for fungal killing in liquid co-cultures24, the spatial restriction during growth on agar plates demonstrates the toxicity of the phenazines of P. aeruginosa towards both yeast and hyphal forms of C. albicans23. These in vitro interactions may reflect the antagonism that is observed between P. aeruginosa and C. albicans (and other fungi) in chronic infections, as evidenced by the increase in fungal growth on host treatment with antibacterial compounds52.
a | Pseudomonas aeruginosa can attach to the surface of Candida albicans hyphae (but not yeast cells) and form biofilms24. Production of phospholipase C24 and phenazines23,94 by P. aeruginosa leads to the death of the fungal filament. b | In the mixed-species biofilm, quorum-sensing molecules that are produced by both P. aeruginosa and C. albicans have roles in autoregulation and interspecies communication9,15,19. Acyl homoserine lactones (AHLs) such as 3-oxo-C12-homoserine lactone produced by P. aeruginosa inhibit the Ras1–cyclic AMP (cAMP)–protein kinase A (PKA) pathway for hyphal growth in C. albicans, thereby inhibiting filamentation of the fungus18. Because yeast cells have increased survival in the presence of P. aeruginosa, the switch to growth as yeast may contribute to the coexistence of both species in mixed infections. c | C. albicans modulates the behaviour of P. aeruginosa through the production of farnesol15, which alters quorum-sensing regulation. Other uncharacterized C. albicans factors increase the production of virulence factors or alter swarming motility and biofilm formation9,23.
C. albicans populations secrete a quorum-sensing molecule called farnesol, which represses hyphal growth despite cues that normally trigger filamentation, such as serum and 37 °C temperatures53. Farnesol acts by repressing elements in the Ras1–cyclic AMP–protein kinase A pathway, which positively regulates hyphal growth18, and effects on other signalling pathways have also been observed. Interestingly, 3-oxo-C12-homoserine lactone, a quorum-sensing molecule produced by P. aeruginosa, has a similar effect on C. albicans9,19, and evidence suggests that it acts by a similar mechanism to farnesol18 (Fig. 3b). Furthermore, other organisms, including Burkholderia cenocepacia 17 and Xanthomonas campestris 22, produce decenoic acids that similarly repress hyphal growth in C. albicans. The finding that C. albicans morphology is influenced by secreted factors that are produced by phylogenetically diverse species opens up three important issues. The first issue is whether the production of bacterial molecules like 3-oxo-C12-homoserine lactone influences the behaviour of co-infecting fungal pathogens in vivo and whether this effect also reflects how these compounds impact human cells. The second issue is whether these molecules affect other fungi and whether these activities can be exploited to attenuate fungal virulence. The third issue is whether other bacterial pathogens or members of the commensal microflora produce molecules with similar effects on C. albicans or other fungi.
C. albicans also affects P. aeruginosa in a range of ways (Fig. 3c). Secreted factors produced by C. albicans inhibit swarming motility in P. aeruginosa, which may lead to enhanced P. aeruginosa biofilm formation on surfaces such as catheters or ventilator apparatus9,54,55. Moreover, farnesol alters the regulation of quorum sensing in P. aeruginosa15. The presence of the fungus may also enhance the production of bacterial virulence factors such as phenazines23.
Interactions between Acinetobacter spp. and fungi. Acinetobacter spp. are Gram-negative, non-fermenting,oxidase-negative bacteria that are ubiquitous in the environment56. The most clinically important species, Acinetobacter baumannii , has evolved to become a highly troublesome pathogen in hospitals worldwide, causing a range of infectious syndromes, including VAP and catheter-related bloodstream infection56,57,58. In health care institutions A. baumannii and C. albicans commonly co-inhabit ecological niches, including urinary and vascular catheters, ventilator tubing and patient wounds10,59,60. In contrast to interactions between fungi and Pseudomonas spp., the first interactions between fungi and Acinetobacter spp. were only recently described16,61,62. Smith et al. reported that Acinetobacter spp., including A. baumannii, exhibited enhanced growth in the presence of Saccharomyces cerevisiae , as a result of ethanol secretion by the yeast62. Furthermore, in the presence of ethanol, Acinetobacter spp. were more resistant to osmotic stress and were also more virulent, as determined by using a Caenorhabditis elegans infection model62. These data may help to explain why Acinetobacter spp. have become so successful in the hospital environment.
In contrast to this synergistic relationship, an antagonistic interaction between the more clinically relevant yeast, C. albicans, and A. baumannii has been described16. It was shown, through the use of a C. elegans polymicrobial infection assay, that A. baumannii is able to inhibit several important virulence determinants of C. albicans, including hyphae and biofilm formation16 (Fig. 4). Interestingly, the inhibitory effect of A. baumannii toward C. albicans attenuated the virulence of the fungus, as determined by reduced killing of C. elegans16. A recent study showed that outer-membrane protein A (OmpA; also known as Omp38) of A. baumannii is essential for the attachment of the bacterium to C. albicans filaments63. Interestingly, this investigation also showed that OmpA is essential for bacterial attachment to mammalian epithelial cells and that apoptotic cell death follows attachment63.
a | When Caenorhabditis elegans is infected with the yeast Candida albicans and then incubated in liquid medium, the yeast cells build up in the gut of the worm and then undergo a morphogenic transition to filamentous cells (green fluorescent cells)16. These filaments penetrate the body of the worm (white arrow), leading to the worm's death. b | Interestingly, when the worm is infected with both C. albicans and the Gram-negative, pathogenic bacterium Acinetobacter baumannii, the number of worms dying with penetrative filamentation is significantly lower. The bacteria inhibit the penetrative C. albicans filamentation in the worm. This bacterial antagonism of the fungus leads to reduced virulence of the fungus toward C. elegans, as determined by the reduced killing that is seen with a mixed infection. The scale bars represent 20 μm. Images reproduced, with permission, from Ref. 16 © (2008) National Academy of Sciences, USA.
Highlighting the complexity of polymicrobial infections, C. albicans is also capable of mounting a counteroffensive against A. baumannii16. When C. albicans cells are allowed to form a quorum in a biofilm environment, the viability of A. baumannii grown in this environment is reduced. As has been shown for other bacteria (Fig. 5), this effect was mediated by farnesol16.
Farnesol inhibits the Pseudomonas spp. quinolone signal (PQS), which is important for quorum sensing in pseudomonal species, and also reduces the production of the pseudomonal virulence factor pyocyanin15. Furthermore, farnesol inhibits the viability of Acinetobacter baumannii in planktonic and biofilm environments16 and also increases the susceptibility of Escherichia coli to antibacterials95. Finally, farnesol is able to interfere with membrane integrity in Staphylococcus aureus, reduce the viability of the bacterium, inhibit its ability to form biofilms and increase its susceptibility to antibacterials20,21,95.
Other interactions between Gram-negative bacteria and fungi. Numerous microbial interactions occur in the human gastrointestinal tract, and the gut is a common site for C. albicans colonization. In addition to the interactions between C. albicans, E. coli and S. marcescens, an interaction between the human gastrointestinal tract pathogen Salmonella enterica subsp. enterica serovar Typhimurium and C. albicans has been reported recently27. The C. elegans polymicrobial infection model was used to study this interaction, as S. Typhimurium and C. albicans both cause a persistent and lethal gut infection in the nematode64,65,66. As seen with other Gram-negative pathogens, S. Typhimurium antagonized C. albicans by inhibiting its growth and the formation of filaments and biofilms. The interaction seemed to be mediated by a heat-stable secretory molecule that was produced from late exponential phase onwards. Mutation in an S. Typhimurium quorum sensor caused no difference in the observed inhibition of C. albicans, and the identity of the active molecule is currently unknown. These data suggest that S. Typhimurium uses aggressive techniques to antagonize commensal gastrointestinal organisms for its pathogenesis towards humans.
Another example of a cross-kingdom interaction mediated by a secreted small molecule is that between Burkholderia cepacia and C. albicans17. The bacterium B. cepacia is another Gram-negative organism that predominantly infects the lungs of patients who are immunocompromized or who have chronic lung disease, including chronic granulomatous disease and cystic fibrosis. Boon and colleagues identified a novel signalling molecule in B. cenocepacia that is a structural homologue of a quorum-sensing molecule and that is known as diffusible signal factor (DSF)17. This molecule inhibited germ tube and filament formation in C. albicans. Similarly to the signalling molecule 3-oxo-C12-homoserine lactone from P. aeruginosa19, DSF from B. cepacia and B. cenocepacia was found to be structurally related to farnesol17. We are only just beginning to appreciate the degree of conservation between the small signalling molecules that are involved in cross-kingdom interactions and their potential importance to polymicrobial infections and human disease.
Another example of virulence enhancement in the setting of a polymicrobial interaction is the interaction between the bacterium Klebsiella aerogenes and the yeast C. neoformans67. This opportunistic fungus can lead to serious disseminated and central nervous system infections in immunocompromized patients68. After in vitro co-cultures on agar, it was observed that colonies of C. neoformans turned brown in the presence of K. aerogenes. This discoloration was due to the presence of melanin, a potent free radical scavenger69. Analysis of the K. aerogenes culture filtrate identified dopamine as a possible substrate that could be used by C. neoformans for melanin production67. These studies and the finding that C. neoformans can use a bacterial melanin precursor compound70 suggest that certain bacteria may augment cryptococcal melanization, protecting it from macrophages.
Interactions between oral streptococci and Candida spp. Diverse arrays of microorganisms inhabit the oral cavity and are responsible for common oral diseases such as denture stomatitis2,71,72. Mixed-species biofilms develop through co-aggregation and are thought to be important for the development of dental plaque and subsequent complications. Oral streptococci, especially Streptococcus gordonii , Streptococcus oralis and Streptococcus sanguinis , adhere well to C. albicans72,73. This adherence, at least for S. gordonii, seems to be mediated through streptococcal cell surface polysaccharide receptors and polypeptide adhesins74,75. Interestingly, these streptococcal species are able to adsorb protein components from human saliva (specifically basic, proline-rich proteins) that promote C. albicans adhesion72. Whether these mechanisms of adhesion have an evolutionary origin and there is some advantage for a mixed bacterial–fungal biofilm is currently unknown. It is plausible that C. albicans has adapted receptors to promote its adhesion to surfaces that are coated with saliva, enabling it to colonize and survive in the oral cavity. Recently, C. albicans adhesins such as Als1 and Als5 were found to be important for adhesion and aggregation with bacterial cells76.
Interactions between staphylococci and Candida spp. Electron microscopy of a Staphylococcus epidermidis – C. albicans biofilm on vascular catheter material showed that the bacterium adhered to both morphological forms of the fungus77. Interestingly, compared with a single-organism biofilm, the mixed biofilm seemed more resistant to antimicrobials such as fluconazole and vancomycin. It was concluded that the extracellular matrix formed by S. epidermidis may protect C. albicans from the antifungal agent. Further work is required to understand the relevance of mixed biofilms for the predisposition of fungi to invasive infection and their susceptibility to antimicrobials and host immune defences.
The effects of the Candida spp. quorum-sensing molecule farnesol on bacteria are not limited to Gram-negative organisms (Fig. 5). Farnesol has also been shown to reduce the viability and biofilm capabilities of S. aureus20,21. This is thought to be mediated by a disruption in cell membrane integrity, as seen by an increase in ethidium bromide uptake and K+ loss in the presence of farnesol. Importantly, the susceptibility of S. aureus to antibiotics increased in the presence of farnesol21, presumably owing to cell membrane damage and greater diffusion of antibiotics to target sites.
Interactions between lactobacilli and Candida spp. Lactobacilli, which normally inhabit mucosal surfaces associated with the intestinal and female reproductive tracts, have been well studied for their potential to protect against pathogens such as C. albicans. The clinical importance of this relationship is highlighted by the complicating effects of systemic antibiotics with activity against lactobacilli, which often lead to vaginitis caused by Candida spp. Furthermore, animal model experiments have shown that in vivo suppression of C. albicans occurs in some cases78,79. Many different mechanisms by which lactobacilli could inhibit the growth and virulence of C. albicans have been proposed, including the production of hydrogen peroxide or the secretion of organic acids28,80,81, and there are many reports of Lactobacillus spp. with uncharacterized activities against the fungus. Studies using cell culture models have led researchers to propose that some Lactobacillus spp. can modulate the virulence of C. albicans through bacterial effects on the host immune response82,83. Physical interactions between Lactobacillus spp. such as Lactobacillus rhamnosus and vaginal or cervical epithelial cells or fungi have been observed, and these interactions may contribute to the attenuation of fungal adherence and invasion78,84,85. The potential for lactobacilli to play a part in protection against fungal infection is exciting and warrants additional research efforts.
Concluding remarks
Medically important interactions between bacteria and fungi are common. These interactions are highly complex, and the type of interaction that occurs often depends on a range of environmental, pathogen and host factors. Developing appropriate in vitro and in vivo models to characterize these interactions and their molecular details is imperative for our understanding of their importance to human disease. Furthermore, exploiting the mechanisms that are used by competing microorganisms to antagonize each other could potentially lead to novel treatment options for problematic human pathogens, and this is now, more than ever, a great necessity.
References
Fleming, A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 10, 226–236 (1929).
Baena-Monroy, T. et al. Candida albicans, Staphylococcus aureus and Streptococcus mutans colonization in patients wearing dental prosthesis. Med. Oral Patol. Oral Cir. Bucal 10, E27–E39 (2005).
Gupta, N., Haque, A., Mukhopadhyay, G., Narayan, R. P. & Prasad, R. Interactions between bacteria and Candida in the burn wound. Burns 31, 375–378 (2005).
Hermann, C., Hermann, J., Munzel, U. & Ruchel, R. Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 42, 619–627 (1999).
Dyess, D. L., Garrison, R. N. & Fry, D. E. Candida sepsis. Implications of polymicrobial blood-borne infection. Arch. Surg. 120, 345–348 (1985).
Verghese, A., Prabhu, K., Diamond, R. D. & Sugar, A. Synchronous bacterial and fungal septicemia. A marker for the critically ill surgical patient. Am. Surg. 54, 276–283 (1988).
Tchekmedyian, N. S. et al. Special studies of the Hickman catheter of a patient with recurrent bacteremia and candidemia. Am. J. Med. Sci. 291, 419–424 (1986).
Bauernfeind, A. et al. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection 15, 270–277 (1987).
McAlester, G., O'Gara, F. & Morrissey, J. P. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J. Med. Microbiol. 57, 563–569 (2008).
Rosenthal, V. D. et al. Device-associated nosocomial infections in 55 intensive care units of 8 developing countries. Ann. Intern. Med. 145, 582–591 (2006).
Azoulay, E. et al. Candida colonization of the respiratory tract and subsequent Pseudomonas ventilator-associated pneumonia. Chest 129, 110–117 (2006). This article shows the importance of bacterial–fungal interactions in the clinical setting.
Adair, C. G. et al. Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med. 25, 1072–1076 (1999).
Kojic, E. M. & Darouiche, R. O. Candida infections of medical devices. Clin. Microbiol. Rev. 17, 255–267 (2004).
Marrie, T. J. & Costerton, J. W. Scanning and transmission electron microscopy of in situ bacterial colonization of intravenous and intraarterial catheters. J. Clin. Microbiol. 19, 687–693 (1984).
Cugini, C. et al. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 65, 896–906 (2007).
Peleg, A. Y. et al. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 105, 14585–14590 (2008). This study demonstrates for the first time the use of a non-mammalian model system to investigate bacterial–fungal interactions.
Boon, C. et al. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2, 27–36 (2008).
Davis-Hanna, A., Piispanen, A. E., Stateva, L. I. & Hogan, D. A. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol. Microbiol. 67, 47–62 (2008).
Hogan, D. A., Vik, A. & Kolter, R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54, 1212–1223 (2004). This paper describes a mechanism, mediated through a quorum-sensing molecule, by which P. aeruginosa inhibits the morphogenic transition of C. albicans from yeast to hyphae.
Inoue, Y. et al. The antibacterial effects of terpene alcohols on Staphylococcus aureus and their mode of action. FEMS Microbiol. Lett. 237, 325–331 (2004).
Jabra-Rizk, M. A., Meiller, T. F., James, C. E. & Shirtliff, M. E. Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob. Agents Chemother. 50, 1463–1469 (2006).
Wang, L. H. et al. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51, 903–912 (2004).
Gibson, J., Sood, A. & Hogan, D. A. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl. Environ. Microbiol. 75, 504–513 (2009).
Hogan, D. A. & Kolter, R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296, 2229–2232 (2002). This article discusses the interactions between two important human pathogens at the molecular level and shows that known pseudomonal virulence factors are important for the antagonistic relationship of Pseudomonas spp. with C. albicans.
Kerr, J. Inhibition of fungal growth by Pseudomonas aeruginosa and Pseudomonas cepacia isolated from patients with cystic fibrosis. J. Infect. 28, 305–310 (1994).
Kerr, J. R. Suppression of fungal growth exhibited by Pseudomonas aeruginosa. J. Clin. Microbiol. 32, 525–527 (1994).
Tampakakis, E., Peleg, A. Y. & Mylonakis, E. The interaction of Candida albicans with an intestinal pathogen; Salmonella enterica serovar Typhimurium. Eukaryot. Cell 8, 732–737 (2009).
Fitzsimmons, N. & Berry, D. R. Inhibition of Candida albicans by Lactobacillus acidophilus: evidence for the involvement of a peroxidase system. Microbios 80, 125–133 (1994).
Buffo, J., Herman, M. A. & Soll, D. R. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia 85, 21–30 (1984).
Klotz, S. A., Chasin, B. S., Powell, B., Gaur, N. K. & Lipke, P. N. Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 59, 401–406 (2007).
Nseir, S. et al. Impact of antifungal treatment on Candida-Pseudomonas interaction: a preliminary retrospective case-control study. Intensive Care Med. 33, 137–142 (2007).
Gudlaugsson, O. et al. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37, 1172–1177 (2003).
Gale, D. & Sandoval, B. Response of mice to the inoculations of both Candida albicans and Escherichia coli. I. The enhancement phenomenon. J. Bacteriol. 73, 616–624 (1957).
Akagawa, G., Abe, S. & Yamaguchi, H. Mortality of Candida albicans-infected mice is facilitated by superinfection of Escherichia coli or administration of its lipopolysaccharide. J. Infect. Dis. 171, 1539–1544 (1995).
Burd, R. S., Raymond, C. S. & Dunn, D. L. Endotoxin promotes synergistic lethality during concurrent Escherichia coli and Candida albicans infection. J. Surg. Res. 52, 537–542 (1992).
Ikeda, T., Suegara, N., Abe, S. & Yamaguchi, H. Efficacy of antibacterial drugs in mice with complex infection by Candida albicans and Escherichia coli. J. Antibiot. (Tokyo) 52, 552–558 (1999).
Klaerner, H. G. et al. Candida albicans and Escherichia coli are synergistic pathogens during experimental microbial peritonitis. J. Surg. Res. 70, 161–165 (1997).
Neely, A. N., Law, E. J. & Holder, I. A. Increased susceptibility to lethal Candida infections in burned mice preinfected with Pseudomonas aeruginosa or pretreated with proteolytic enzymes. Infect. Immun. 52, 200–204 (1986).
Roux., D. et al. Candida albicans impairs macrophage function and facilitates Pseudomonas aeruginosa pneumonia in rat. Crit. Care Med. 37, 1062–1067 (2009). This study demonstrates the effect of C. albicans colonization of the airway on pseudomonal pneumonia in rats, showing that pre-colonization with live C. albicans increases pseudomonal pneumonia and that this may be partly due to impaired macrophage function.
Xu, X. L. et al. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 4, 28–39 (2008). This work finds that bacterial peptidoglycan products, which are present in human blood, can stimulate an important virulence determinant in C. albicans : hyphal growth.
Lo, H. J. et al. Nonfilamentous C. albicans mutants are avirulent. Cell 90, 939–949 (1997).
Carlson, E. Enhancement by Candida albicans of Staphylococcus aureus, Serratia marcescens, and Streptococcus faecalis in the establishment of infection in mice. Infect. Immun. 39, 193–197 (1983).
Carlson, E. & Johnson, G. Protection by Candida albicans of Staphylococcus aureus in the establishment of dual infection in mice. Infect. Immun. 50, 655–659 (1985).
Carlson, E. Effect of strain of Staphylococcus aureus on synergism with Candida albicans resulting in mouse mortality and morbidity. Infect. Immun. 42, 285–292 (1983).
Allard, J. B. et al. Th2 allergic immune response to inhaled fungal antigens is modulated by TLR-4-independent bacterial products. Eur. J. Immunol. 39, 776–788 (2009). This article describes the use of a mammalian model system to investigate the immunological responses to lung exposure to P. aeruginosa and C. albicans antigens.
Braun-Fahrlander, C. et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N. Engl. J. Med. 347, 869–877 (2002).
Garn, H. & Renz, H. Epidemiological and immunological evidence for the hygiene hypothesis. Immunobiology 212, 441–452 (2007).
Bach, J. F. The effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 347, 911–920 (2002).
de Macedo, J. L. & Santos, J. B. Bacterial and fungal colonization of burn wounds. Mem. Inst. Oswaldo Cruz 100, 535–539 (2005).
Hughes, W. T. & Kim, H. K. Mycoflora in cystic fibrosis: some ecologic aspects of Pseudomonas aeruginosa and Candida albicans. Mycopathol. Mycol. Appl. 50, 261–269 (1973).
Hockey, L. J. et al. Detection of fungemia obscured by concomitant bacteremia: in vitro and in vivo studies. J. Clin. Microbiol. 16, 1080–1085 (1982).
Burns, J. L. et al. Effect of chronic intermittent administration of inhaled tobramycin on respiratory microbial flora in patients with cystic fibrosis. J. Infect. Dis. 179, 1190–1196 (1999).
Hornby, J. M. et al. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67, 2982–2992 (2001).
Caiazza, N. C., Merritt, J. H., Brothers, K. M. & O'Toole, G. A. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 3603–3612 (2007).
Shrout, J. D. et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62, 1264–1277 (2006).
Peleg, A. Y., Seifert, H. & Paterson, D. L. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 (2008).
Dijkshoorn, L., Nemec, A. & Seifert, H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nature Rev. Microbiol. 5, 939–951 (2007).
Munoz-Price, L. S. & Weinstein, R. A. Acinetobacter infection. N. Engl. J. Med. 358, 1271–1281 (2008).
Chim., H., Tan, B. H. & Song, C. Five-year review of infections in a burn intensive care unit: high incidence of Acinetobacter baumannii in a tropical climate. Burns 33, 1008–1014 (2007).
Richards, M. J., Edwards, J. R., Culver, D. H. & Gaynes, R. P. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit. Care Med. 27, 887–892 (1999).
Liu, C. H. et al. Study of the antifungal activity of Acinetobacter baumannii LCH001 in vitro and identification of its antifungal components. Appl. Microbiol. Biotechnol. 76, 459–466 (2007).
Smith, M. G., Des Etages, S. G. & Snyder, M. Microbial synergy via an ethanol-triggered pathway. Mol. Cell Biol. 24, 3874–3884 (2004).
Gaddy, J. A., Tomaras, A. P. & Actis, L. A. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77, 3150–3160 (2009). This study shows the importance of a known A. baumannii virulence factor, OmpA, for the interaction of this bacterium with both C. albicans and human alveolar epithelial cells.
Aballay, A., Yorgey, P. & Ausubel, F. M. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10, 1539–1542 (2000).
Breger, J. et al. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 3, e18 (2007).
Labrousse, A., Chauvet, S., Couillault, C., Kurz, C. L. & Ewbank, J. J. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 10, 1543–1545 (2000).
Frases, S., Chaskes, S., Dadachova, E. & Casadevall, A. Induction by Klebsiella aerogenes of a melanin-like pigment in Cryptococcus neoformans. Appl. Environ. Microbiol. 72, 1542–1550 (2006).
Pukkila-Worley, R. & Mylonakis, E. Epidemiology and management of cryptococcal meningitis: developments and challenges. Expert Opin. Pharmacother. 9, 551–560 (2008).
Nosanchuk, J. D. & Casadevall, A. The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5, 203–223 (2003).
Frases, S., Salazar, A., Dadachova, E. & Casadevall, A. Cryptococcus neoformans can utilize the bacterial melanin precursor homogentisic acid for fungal melanogenesis. Appl. Environ. Microbiol. 73, 615–621 (2007).
Kroes, I., Lepp, P. W. & Relman, D. A. Bacterial diversity within the human subgingival crevice. Proc. Natl Acad. Sci. USA 96, 14547–14552 (1999).
O'Sullivan, J. M., Jenkinson, H. F. & Cannon, R. D. Adhesion of Candida albicans to oral streptococci is promoted by selective adsorption of salivary proteins to the streptococcal cell surface. Microbiol. 146, 41–48 (2000).
Jenkinson, H. F., Lala, H. C. & Shepherd, M. G. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect. Immun. 58, 1429–1436 (1990).
Holmes, A. R., Gopal, P. K. & Jenkinson, H. F. Adherence of Candida albicans to a cell surface polysaccharide receptor on Streptococcus gordonii. Infect. Immun. 63, 1827–1834 (1995).
Holmes, A. R., McNab, R. & Jenkinson, H. F. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64, 4680–4685 (1996). This work uncovers the complex physical interactions that can occur between bacteria and fungi.
Klotz, S. A. et al. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med. Mycol. 45, 363–370 (2007).
Adam, B., Baillie, G. S. & Douglas, L. J. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J. Med. Microbiol. 51, 344–349 (2002).
Pope, L. M., Cole, G. T., Guentzel, M. N. & Berry, L. J. Systemic and gastrointestinal candidiasis of infant mice after intragastric challenge. Infect. Immun. 25, 702–707 (1979).
Wagner, R. D. et al. Biotherapeutic effects of probiotic bacteria on candidiasis in immunodeficient mice. Infect. Immun. 65, 4165–4172 (1997).
Noverr, M. C. & Huffnagle, G. B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 72, 6206–6210 (2004).
Hoberg, K. A., Cihlar, R. L. & Calderone, R. A. Inhibitory effect of cerulenin and sodium butyrate on germination of Candida albicans. Antimicrob. Agents Chemother. 24, 401–408 (1983).
Martinez, R. C. et al. Effect of Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 on the ability of Candida albicans to infect cells and induce inflammation. Microbiol. Immunol. 53, 487–495 (2009).
Spear, G. T., Zariffard, M. R., Cohen, M. H. & Sha, B. E. Vaginal IL-8 levels are positively associated with Candida albicans and inversely with lactobacilli in HIV-infected women. J. Reprod. Immunol. 78, 76–79 (2008). A clinical study that provides the basis for future work on bacterial–fungal interactions that can be mediated by the host immune response.
Coudeyras, S., Jugie, G., Vermerie, M. & Forestier, C. Adhesion of human probiotic Lactobacillus rhamnosus to cervical and vaginal cells and interaction with vaginosis-associated pathogens. Infect. Dis. Obstet. Gynecol. 2008, 549640 (2008).
Mastromarino, P. et al. Characterization and selection of vaginal Lactobacillus strains for the preparation of vaginal tablets. J. Appl. Microbiol. 93, 884–893 (2002).
Shen, S., Samaranayake, L. P. & Yip, H. K. Coaggregation profiles of the microflora from root surface caries lesions. Arch. Oral Biol. 50, 23–32 (2005).
Grillot, R., Portmann-Coffin, V. & Ambroise-Thomas, P. Growth inhibition of pathogenic yeasts by Pseudomonas aeruginosa in vitro: clinical implications in blood cultures. Mycoses 37, 343–347 (1994).
Kennedy, M. J., Rogers, A. L. & Yancey, R. J. Jr. An anaerobic continuous-flow culture model of interactions between intestinal microflora and Candida albicans. Mycopathologia 103, 125–134 (1988).
O'May, G. A., Reynolds, N. & Macfarlane, G. T. Effect of pH on an in vitro model of gastric microbiota in enteral nutrition patients. Appl. Environ. Microbiol. 71, 4777–4783 (2005).
Kaleli, I., Cevahir, N., Demir, M., Yildirim, U. & Sahin, R. Anticandidal activity of Pseudomonas aeruginosa strains isolated from clinical specimens. Mycoses 50, 74–78 (2007).
Fidel, P. L. Jr, Cutright, J. L., Tait, L. & Sobel, J. D. A murine model of Candida glabrata vaginitis. J. Infect. Dis. 173, 425–431 (1996).
Sibley, C. D. et al. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 4, e1000184 (2008).
Ocana, V. S. & Nader-Macias, M. E. Vaginal lactobacilli: self- and co-aggregating ability. Br. J. Biomed. Sci. 59, 183–190 (2002).
Kerr, J. R. et al. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 52, 385–387 (1999).
Brehm-Stecher, B. F. & Johnson, E. A. Sensitization of Staphylococcus aureus and Escherichia coli to antibiotics by the sesquiterpenoids nerolidol, farnesol, bisabolol, and apritone. Antimicrob. Agents Chemother. 47, 3357–3360 (2003).
Acknowledgements
A.Y.P. is funded by a Massachusetts General Hospital ECOR Fund for Medical Discovery Award.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
Salmonella enterica subsp. enterica serovar Typhimurium
FURTHER INFORMATION
Glossary
- Gastrointestinal mucositis
-
Inflammation of the mucosal lining of the gastrointestinal tract (which extends from the oral cavity through to the rectum), often leading to increased microbial transit through the gastrointestinal wall.
- Commensal organism
-
A microorganism that resides on or in the host without causing disease.
- Candidiasis
-
Infection with a Candida species.
- Biofilm
-
A complex community of microorganisms that are often attached to a surface and are surrounded by extracellular matrix.
- Quorum sensing
-
Communication between neighbouring organisms through secreted signalling molecules that allows populations to sense organism density and alter gene expression.
- Phenazine
-
A secreted secondary metabolite and virulence factor of P. aeruginosa.
- Probiotic
-
A microorganism that confers a health benefit to the host.
- Endotoxin
-
A toxin that is part of the structure of the bacterium rather than being secreted. In Gram-negative bacteria, it is most commonly lipopolysaccharide in the outer cell membrane.
- Peritoneal cavity
-
The space in the abdomen that is lined by visceral and parietal peritoneum.
- Atopic disease
-
A disease associated with an allergy (that is, mediated by immunoglobulin E), such as asthma, eczema and hay fever.
Rights and permissions
About this article
Cite this article
Peleg, A., Hogan, D. & Mylonakis, E. Medically important bacterial–fungal interactions. Nat Rev Microbiol 8, 340–349 (2010). https://doi.org/10.1038/nrmicro2313
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2313
This article is cited by
-
Characterization of microbial communities in urban subway: connotation for indoor environment quality and public health
Air Quality, Atmosphere & Health (2024)
-
Characterization of microbial communities during Grifola frondosa (maitake) wood log cultivation
Journal of Wood Science (2023)
-
Bacterial–fungal interactions promote parallel evolution of global transcriptional regulators in a widespread Staphylococcus species
The ISME Journal (2023)
-
Impact of diet and host genetics on the murine intestinal mycobiome
Nature Communications (2023)
-
Basidiomycota species in Drosophila gut are associated with host fat metabolism
Scientific Reports (2023)