Abstract
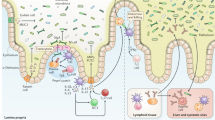
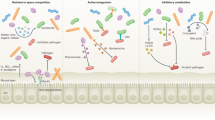
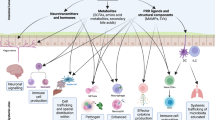
The mammalian intestine is home to dense and complex indigenous bacterial communities. Most of these bacteria establish beneficial symbiotic relationships with their hosts, making important contributions to host metabolism and digestive efficiency. The vast numbers of intestinal bacteria and their proximity to host tissues raise the question of how symbiotic host–bacterial relationships are established without eliciting potentially harmful immune responses. In light of the varied ways in which pathogenic bacteria manipulate host immunity, this Opinion article explores the role of immune suppression, subversion and evasion in the establishment of symbiotic host–bacterial associations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Xu, J. & Gordon, J. I. Inaugural Article: Honor thy symbionts. Proc. Natl Acad. Sci. USA 100, 10452–10459 (2003).
Zhang, T. et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4, e3 (2006).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
Dethlefsen, L., McFall-Ngai, M. & Relman, D. A. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449, 811–818 (2007).
Darveau, R. P., McFall-Ngai, M., Ruby, E., Miller, S. & Mangan, D. Host tissues may actively respond to beneficial microbes. ASM News 186–191 (2003).
Xavier, R. J. & Podolsky, D. K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448, 427–434 (2007).
Peterson, D. A., Frank, D. N., Pace, N. R. & Gordon, J. I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3, 417–427 (2008).
Suau, A. et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65, 4799–4807 (1999).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Palmer, C., Bik, E. M., Digiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007).
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006).
Ley, R. E. et al. Evolution of mammals and their gut microbes. Science 320, 1647–1651 (2008).
Ley, R. E., Lozupone, C. A., Hamady, M., Knight, R. & Gordon, J. I. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Rev. Microbiol. 6, 776–788 (2008).
Wostmann, B. S., Larkin, C., Moriarty, A. & Bruckner-Kardoss, E. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab. Anim. Sci. 33, 46–50 (1983).
McFall-Ngai, M. Adaptive immunity: care for the community. Nature 445, 153 (2007).
Flint, H. J., Bayer, E. A., Rincon, M. T., Lamed, R. & White, B. A. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev. Microbiol. 6, 121–131 (2008).
Sonnenburg, J. L. et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005).
Martens, E. C., Chiang, H. C. & Gordon, J. I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008).
Lozupone, C. A. et al. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc. Natl Acad. Sci. USA 105, 15076–15081 (2008).
Stecher, B. et al. Comparison of Salmonella enterica serovar Typhimurium colitis in germfree mice and mice pretreated with streptomycin. Infect. Immun. 73, 3228–3241 (2005).
Stecher, B. et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5, 2177–2189 (2007).
Sansonetti, P. J. War and peace at mucosal surfaces. Nature Rev. Immunol. 4, 953–964 (2004).
Xu, J. et al. A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003).
Macpherson, A. J. et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000).
Macpherson, A. J., Geuking, M. B. & McCoy, K. D. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology 115, 153–162 (2005).
Neish, A. S. Molecular aspects of intestinal epithelial cell–bacterial interactions that determine the development of intestinal inflammation. Inflamm. Bowel Dis. 10, 159–168 (2004).
Johansson, M. E. et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA 105, 15064–15069 (2008).
Van der Sluis, M. et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131, 117–129 (2006).
Velcich, A. et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295, 1726–1729 (2002).
Mukherjee, S., Vaishnava, S. & Hooper, L. V. Multi-layered regulation of intestinal antimicrobial defense. Cell. Mol. Life Sci. 65, 3019–3027 (2008).
Putsep, K. et al. Germ-free and colonized mice generate the same products from enteric prodefensins. J. Biol. Chem. 275, 40478–40482 (2000).
Hooper, L. V., Stappenbeck, T. S., Hong, C. V. & Gordon, J. I. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature Immunol. 4, 269–273 (2003).
Cash, H. L., Whitham, C. V., Behrendt, C. L. & Hooper, L. V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Brandl, K., Plitas, G., Schnabl, B., Dematteo, R. P. & Pamer, E. G. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 204, 1891–1900 (2007).
Kobayashi, K. S. et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731–734 (2005).
Vaishnava, S., Behrendt, C. L., Ismail, A. S., Eckmann, L. & Hooper, L. V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host–microbial interface. Proc. Natl Acad. Sci. USA 105, 20858–20863 (2008).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Macpherson, A. J. & Uhr, T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303, 1662–1665 (2004).
Fagarasan, S. & Honjo, T. Intestinal IgA synthesis: regulation of front-line body defences. Nature Rev. Immunol. 3, 63–72 (2003).
Andoh, A., Fujiyama, Y., Bamba, T. & Hosoda, S. Differential cytokine regulation of complement C3, C4, and factor B synthesis in human intestinal epithelial cell line, Caco-2. J. Immunol. 151, 4239–4247 (1993).
Pasquier, B. et al. Identification of FcαRI as an inhibitory receptor that controls inflammation: dual role of FcRγ ITAM. Immunity 22, 31–42 (2005).
Rescigno, M. et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nature Immunol. 2, 361–367 (2001).
Guerry, P. Campylobacter flagella: not just for motility. Trends Microbiol. 15, 456–461 (2007).
Zhou, D. & Galan, J. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3, 1293–1298 (2001).
Galan, J. E. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20, 263–271 (1996).
Shi, Y., Cromie, M. J., Hsu, F. F., Turk, J. & Groisman, E. A. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53, 229–241 (2004).
Groisman, E. A., Parra-Lopez, C., Salcedo, M., Lipps, C. J. & Heffron, F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl Acad. Sci. USA 89, 11939–11943 (1992).
Guerry, P. & Szymanski, C. M. Campylobacter sugars sticking out. Trends Microbiol. 16, 428–435 (2008).
Peterson, D. A., McNulty, N. P., Guruge, J. L. & Gordon, J. I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Coyne, M. J., Reinap, B., Lee, M. M. & Comstock, L. E. Human symbionts use a host-like pathway for surface fucosylation. Science 307, 1778–1781 (2005).
Comstock, L. E. & Coyne, M. J. Bacteroides thetaiotaomicron: a dynamic, niche-adapted human symbiont. Bioessays 25, 926–929 (2003).
Krinos, C. M. et al. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414, 555–558 (2001).
Kuwahara, T. et al. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl Acad. Sci. USA 101, 14919–14924 (2004).
Cerdeno-Tarraga, A. M. et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307, 1463–1465 (2005).
Artis, D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nature Rev. Immunol. 8, 411–420 (2008).
Abreu, M. T. et al. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J. Immunol. 167, 1609–1616 (2001).
Melmed, G. et al. Human intestinal epithelial cells are broadly unresponsive to Toll-like receptor 2-dependent bacterial ligands: implications for host–microbial interactions in the gut. J. Immunol. 170, 1406–1415 (2003).
Ortega-Cava, C. F. et al. Strategic compartmentalization of Toll-like receptor 4 in the mouse gut. J. Immunol. 170, 3977–3985 (2003).
Gewirtz, A. T., Navas, T. A., Lyons, S., Godowski, P. J. & Madara, J. L. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167, 1882–1885 (2001).
Nenci, A. et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature 446, 557–561 (2007).
Zaph, C. et al. Epithelial-cell-intrinsic IKK-β expression regulates intestinal immune homeostasis. Nature 446, 552–556 (2007).
Brandl, K. et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807 (2008).
Neish, A. S. et al. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science 289, 1560–1563 (2000).
Kelly, D. et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nature Immunol. 5, 104–112 (2004).
Bates, J. M., Akerlund, J., Mittge, E. & Guillemin, K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382 (2007).
Vaishnava, S. & Hooper, L. V. Alkaline phosphatase: keeping the peace at the gut epithelial surface. Cell Host Microbe 2, 365–367 (2007).
Swidsinski, A., Weber, J., Loening-Baucke, V., Hale, L. P. & Lochs, H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43, 3380–3389 (2005).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
Hugot, J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Wehkamp, J. et al. Reduced Paneth cell α-defensins in ileal Crohn's disease. Proc. Natl Acad. Sci. USA 102, 18129–18134 (2005).
Macpherson, A. J. & Slack, E. The functional interactions of commensal bacteria with intestinal secretory IgA. Curr. Opin. Gastroenterol. 23, 673–678 (2007).
Suzuki, K. et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl Acad. Sci. USA 101, 1981–1986 (2004).
Deng, W. et al. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185, 2330–2337 (2003).
Wei, J. et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71, 2775–2786 (2003).
Acknowledgements
The author thanks the students and colleagues from her laboratory for the many discussions that contributed to the ideas in this manuscript. Work in the author's laboratory is supported by the Howard Hughes Medical Institute, the National Institutes of Health (DK070855), the Burroughs Wellcome Foundation (New Investigators in the Pathogenesis of Infectious Diseases Award) and the Crohn's and Colitis Foundation.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Hooper, L. Do symbiotic bacteria subvert host immunity?. Nat Rev Microbiol 7, 367–374 (2009). https://doi.org/10.1038/nrmicro2114
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2114
This article is cited by
-
The impact of mass drug administration of antibiotics on the gut microbiota of target populations
Infectious Diseases of Poverty (2022)
-
The Connection Between Physical Exercise and Gut Microbiota: Implications for Competitive Sports Athletes
Sports Medicine (2022)
-
Joint contributions of the gut microbiota and host genetics to feed efficiency in chickens
Microbiome (2021)
-
Gut microbiota and the human gut physiological changes
Annals of Microbiology (2020)
-
Glycan utilisation system in Bacteroides and Bifidobacteria and their roles in gut stability and health
Applied Microbiology and Biotechnology (2019)