Key Points
-
Our ability to reconstruct genome-scale metabolic networks in microbial cells from genomic and high-throughput data has grown substantially in recent years. There are currently more than 25 genome-scale metabolic reconstructions of microbial cells, and 6–10 more are being produced each year.
-
An increasing number of research groups around the world are working on genome-scale reconstructions of metabolism in their target organism.
-
There is no single source that practitioners can access to learn about and understand the reconstruction process.
-
This Review details the data flows and work flows that underlie the reconstruction process and thus provides a basis for newcomers in the field.
-
Biological network reconstructions continue to grow in scope and are expected to include transcriptional regulation and protein synthesis over the next few years. Expansion in scope will probably also include small RNAs and two-component signalling networks.
-
Genome-scale reconstructions are a common denominator in systems biology of microorganisms and are reaching an advanced stage of development, which indicates that systems analysis of microbial functions and phenotypes will progress in the years to come.
Abstract
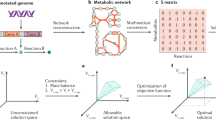
Systems analysis of metabolic and growth functions in microbial organisms is rapidly developing and maturing. Such studies are enabled by reconstruction, at the genomic scale, of the biochemical reaction networks that underlie cellular processes. The network reconstruction process is organism specific and is based on an annotated genome sequence, high-throughput network-wide data sets and bibliomic data on the detailed properties of individual network components. Here we describe the process that is currently used to achieve comprehensive network reconstructions and discuss how these reconstructions are curated and validated. This Review should aid the growing number of researchers who are carrying out reconstructions for particular target organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Reed, J. L., Famili, I., Thiele, I. & Palsson, B. O. Towards multidimensional genome annotation. Nature Rev. Genet. 7, 130–141 (2006). A review of the conceptual basis for network reconstruction.
Price, N. D., Reed, J. L. & Palsson, B. O. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nature Rev. Microbiol. 2, 886–897 (2004). A comprehensive and succinct review of COBRA methods.
Becker, S. A. et al. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA toolbox. Nature Protoc. 2, 727–738 (2007).
Feist, A. M. & Palsson, B. O. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nature Biotechnol. 26, 659–667 (2008). A review of the history of applications of the genome-scale E. coli metabolic reconstruction.
Papoutsakis, E. T. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol.Bioeng. 26, 174–187 (1984).
Papoutsakis, E. & Meyer, C. Fermentation equations for propionic-acid bacteria and production of assorted oxychemicals from various sugars. Biotechnol. Bioeng. 27, 67–80 (1985).
Papoutsakis, E. & Meyer, C. Equations and calculations of product yields and preferred pathways for butanediol and mixed-acid fermentations. Biotechnol. Bioeng. 27, 50–66 (1985).
Majewski, R. A. & Domach, M. M. Simple constrained optimization view of acetate overflow in E. coli. Biotechnol. Bioeng. 35, 732–738 (1990).
Varma, A., Boesch, B. W. & Palsson, B. O. Stoichiometric interpretation of Escherichia coli glucose catabolism under various oxygenation rates. Appl. Environ. Microbiol. 59, 2465–2473 (1993).
Varma, A., Boesch, B. W. & Palsson, B. O. Biochemical production capabilities of Escherichia coli. Biotechnol. Bioeng. 42, 59–73 (1993).
Karp, P. D. et al. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 35, 7577–7590 (2007).
Christie, K. R. et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32, D311–D314 (2004).
Guldener, U. et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 33, D364–D368 (2005).
Maglott, D., Ostell, J., Pruitt, K. D. & Tatusova, T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 35, D26–D31 (2007).
Peterson, J. D., Umayam, L. A., Dickinson, T., Hickey, E. K. & White, O. The Comprehensive Microbial Resource. Nucleic Acids Res. 29, 123–125 (2001).
Stoesser, G., Tuli, M. A., Lopez, R. & Sterk, P. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 27, 18–24 (1999).
Markowitz, V. M. et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34, D344–D348 (2006).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Schomburg, I. et al. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 32, D431–D433 (2004).
Krieger, C. J. et al. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 32, D438–D442 (2004).
DeJongh, M. et al. Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics 8, 139 (2007). An innovative approach for semi-automatic reconstruction of genome-scale metabolic networks that combines automated genome annotation tools with model-based gap filling.
Ren, Q., Chen, K. & Paulsen, I. T. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 35, D274–D279 (2007).
Neidhardt, F. C. (ed.) Escherichia coli and Salmonella: Cellular and Molecular Biology (ASM Press, Washington DC, 1996).
Dickinson, J. R. & Schweizer, M. (eds) The Metabolism and Molecular Physiology of Saccharomyces cerevisiae 2nd edn (Taylor & Francis, London; Philadelphia, 2004).
Marre, R. et al. (eds) Legionella (ASM Press, Washington DC, 2001).
Mobley, H. L. T., Mendz, G. L. & Hazell, S. L. Helicobacter pylori (ASM Press, Washington DC, 2001).
Huh, W. K. et al. Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003).
Janssen, P., Goldovsky, L., Kunin, V., Darzentas, N. & Ouzounis, C. A. Genome coverage, literally speaking. The challenge of annotating 200 genomes with 4 million publications. EMBO Rep. 6, 397–399 (2005).
Reed, J. L. & Palsson, B. O. Thirteen years of building constraint-based in silico models of Escherichia coli. J. Bacteriol. 185, 2692–2699 (2003).
Feist, A. M. et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 3, 121 (2007).
Notebaart, R. A., van Enckevort, F. H., Francke, C., Siezen, R. J. & Teusink, B. Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics 7, 296 (2006).
Borodina, I. & Nielsen, J. From genomes to in silico cells via metabolic networks. Curr. Opin. Biotechnol. 16, 350–355 (2005).
Lee, S. Y. et al. Systems-level analysis of genome-scale in silico metabolic models using MetaFluxNet. Biotechnol. Bioprocess Eng. 10, 425–431 (2005).
Klamt, S., Saez-Rodriguez, J. & Gilles, E. D. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Syst. Biol. 1, 2 (2007).
Barabasi, A. L. & Oltvai, Z. N. Network biology: understanding the cell's functional organization. Nature Rev. Genet. 5, 101–113 (2004).
Kwast, K. E. et al. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184, 250–265 (2002).
Neidhardt, F. C. & Umbarger, H. E. in Escherichia coli and Salmonella: Cellular and Molecular Biology (ed. Neidhardt, F. C.) 13–16 (ASM Press, Washington DC, 1996).
Joyce, A. R. et al. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J. Bacteriol. 188, 8259–8271 (2006).
Cherry, J. M. et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 26, 73–79 (1998).
Schuetz, R., Kuepfer, L. & Sauer, U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 3, 119 (2007).
Knorr, A. L., Jain, R. & Srivastava, R. Bayesian-based selection of metabolic objective functions. Bioinformatics 23, 351–357 (2007).
Breitling, R., Vitkup, D. & Barrett, M. P. New surveyor tools for charting microbial metabolic maps. Nature Rev. Microbiol. 6, 156–161 (2008). A review of available computational tools that can improve and expand biological network reconstructions.
Varma, A. & Palsson, B. O. Parametric sensitivity of stoichiometric flux balance models applied to wild-type Escherichia coli metabolism. Biotechnol. Bioeng. 45, 69–79 (1995).
Reed, J. L. et al. Systems approach to refining genome annotation. Proc. Natl Acad. Sci. USA 103, 17480–17484 (2006). The first demonstration of the gap-filling process: a network-guided discovery process.
Roussel, M. R. & Zhu, R. Stochastic kinetics description of a simple transcription model. Bull. Math. Biol. 68, 1681–1713 (2006).
Mehra, A. & Hatzimanikatis, V. An algorithmic framework for genome-wide modeling and analysis of translation networks. Biophys. J. 90, 1136–1146 (2006).
Weitzke, E. L. & Ortoleva, P. J. Simulating cellular dynamics through a coupled transcription, translation, metabolic model. Comput. Biol. Chem. 27, 469–480 (2003).
Allen, T. E. & Palsson, B. O. Sequenced-based analysis of metabolic demands for protein synthesis in prokaryotes. J. Theor. Biol. 220, 1–18 (2003).
Drew, D. A. A mathematical model for prokaryotic protein synthesis. Bull. Math. Biol. 63, 329–351 (2001).
Karp, P. D. et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 33, 6083–6089 (2005).
Sprinzl, M. & Vassilenko, K. S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 33, D139–D140 (2005).
Neidhardt, F. C., Ingraham, J. L. & Schaechter, M. Physiology of The Bacterial Cell: a Molecular Approach (Sinauer Associates, Sunderland, Massachusetts, 1990).
Cho, B. K., Knight, E. M., Barrett, C. L. & Palsson, B. Ø. Genome-wide analysis of Fis binding in Escherichia coli indicates a causative role for A-/AT-tracts. Genome Res. 18, 900–910 (2008).
Dion, M. F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007).
Ren, B. et al. Genome-wide location and function of DNA binding proteins. Science 290, 2306–2309 (2000).
Harbison, C. T. et al. Transcriptional regulatory code of a eukaryotic genome. Nature 431, 99–104 (2004).
Buck, M. J. & Lieb, J. D. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nature Genet. 38, 1446–1451 (2006).
Mukherjee, S. et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nature Genet. 36, 1331–1339 (2004).
Maerkl, S. J. & Quake, S. R. A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007).
Liu, X., Lee, C. K., Granek, J. A., Clarke, N. D. & Lieb, J. D. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 16, 1517–1528 (2006).
Covert, M. W., Knight, E. M., Reed, J. L., Herrgard, M. J. & Palsson, B. O. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429, 92–96 (2004). This study shows the value of literature-based reconstruction of TRNs for well-studied organisms, as well as the integration of metabolic-network and TRN models.
Hu, Z., Killion, P. J. & Iyer, V. R. Genetic reconstruction of a functional transcriptional regulatory network. Nature Genet. 39, 683–687 (2007).
Chua, G. et al. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl Acad. Sci. USA 103, 12045–12050 (2006).
Faith, J. J. et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5, e8 (2007).
Segal, E. et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature Genet. 34, 166–176 (2003).
Workman, C. T. et al. A systems approach to mapping DNA damage response pathways. Science 312, 1054–1059 (2006). References 65 and 67 present alternative statistical approaches for mapping TRNs from large-scale experimental data sets (gene expression or ChIP–chip) obtained for well- characterized model organisms.
Zeitlinger, J. et al. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113, 395–404 (2003).
Kim, J. B. et al. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science 316, 1481–1484 (2007).
Robertson, G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nature Methods 4, 651–657 (2007). This study presents a comprehensive approach to the building of predictive models of large-scale TRNs for even poorly understood organisms using a low number of genome-scale experiments.
Bonneau, R. et al. A predictive model for transcriptional control of physiology in a free living cell. Cell 131, 1354–1365 (2007).
Perez-Rueda, E. & Collado-Vides, J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 28, 1838–1847 (2000).
Price, M. N., Dehal, P. S. & Arkin, A. P. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput. Biol. 3, 1739–1750 (2007).
Salgado, H. et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 34, D394–D397 (2006).
Shmulevich, I., Dougherty, E. R., Kim, S. & Zhang, W. Probabilistic Boolean Networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics 18, 261–274 (2002).
Segal, E., Raveh-Sadka, T., Schroeder, M., Unnerstall, U. & Gaul, U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature 451, 535–540 (2008).
Herrgard, M. J., Lee, B. S., Portnoy, V. & Palsson, B. O. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res. 16, 627–635 (2006).
Gianchandani, E. P., Papin, J. A., Price, N. D., Joyce, A. R. & Palsson, B. O. Matrix formalism to describe functional states of transcriptional regulatory systems. PLoS Comput. Biol. 2, e101 (2006).
Kao, K. C. et al. Transcriptome-based determination of multiple transcription regulator activities in Escherichia coli by using network component analysis. Proc. Natl Acad. Sci. USA 101, 641–646 (2004).
Yamamoto, K. et al. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280, 1448–1456 (2005).
Skerker, J. M., Prasol, M. S., Perchuk, B. S., Biondi, E. G. & Laub, M. T. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3, e334 (2005). Combinations of systematic in vivo and in vitro profiling approaches, such as those used in this work, will be needed to decipher the connectivity and function of bacterial two-component systems.
Seshasayee, A. S., Bertone, P., Fraser, G. M. & Luscombe, N. M. Transcriptional regulatory networks in bacteria: from input signals to output responses. Curr. Opin. Microbiol. 9, 511–519 (2006).
Vogel, J. & Wagner, E. G. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 10, 262–270 (2007).
Romby, P., Vandenesch, F. & Wagner, E. G. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 9, 229–236 (2006).
Vogel, J. & Sharma, C. M. How to find small non-coding RNAs in bacteria. Biol. Chem. 386, 1219–1238 (2005).
Altuvia, S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 10, 257–261 (2007).
Shimoni, Y. et al. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol. Syst. Biol. 3, 138 (2007).
Lee, J. M., Gianchandani, E. P., Eddy, J. A. & Papin, J. A. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput. Biol. 4, e1000086 (2008).
Palsson, B. O. Two-dimensional annotation of genomes. Nature Biotechnol. 22, 1218–1219 (2004).
Cakir, T. et al. Flux balance analysis of a genome-scale yeast model constrained by exometabolomic data allows metabolic system identification of genetically different strains. Biotechnol. Prog. 23, 320–326 (2007).
Vemuri, G. N., Eiteman, M. A., McEwen, J. E., Olsson, L. & Nielsen, J. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 104, 2402–2407 (2007).
Pramanik, J. & Keasling, J. D. Stoichiometric model of Escherichia coli metabolism: incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol. Bioeng. 56, 398–421 (1997).
Pramanik, J. & Keasling, J. D. Effect of Escherichia coli biomass composition on central metabolic fluxes predicted by a stoichiometric model. Biotechnol. Bioeng. 60, 230–238 (1998).
Segre, D., Vitkup, D. & Church, G. M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl Acad. Sci. USA 99, 15112–15117 (2002).
Shlomi, T., Berkman, O. & Ruppin, E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc. Natl Acad. Sci. USA 102, 7695–7700 (2005).
Feist, A. M., Scholten, J. C. M., Palsson, B. O., Brockman, F. J. & Ideker, T. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol. Syst. Biol. 2, 1–14 (2006).
Pharkya, P., Burgard, A. P. & Maranas, C. D. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 14, 2367–2376 (2004).
Harrison, R., Papp, B., Pal, C., Oliver, S. G. & Delneri, D. Plasticity of genetic interactions in metabolic networks of yeast. Proc. Natl Acad. Sci. USA 104, 2307–2312 (2007). A combination of genome-scale modelling and experimentation was used to study condition-dependent genetic interactions and identify novel alternative pathways in yeast.
Almaas, E., Kovacs, B., Vicsek, T., Oltvai, Z. N. & Barabasi, A. L. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature 427, 839–843 (2004).
Burgard, A. P., Vaidyaraman, S. & Maranas, C. D. Minimal reaction sets for Escherichia coli metabolism under different growth requirements and uptake environments. Biotechnol. Prog. 17, 791–797 (2001).
Mahadevan, R. & Schilling, C. H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 5, 264–276 (2003).
Hucka, M. et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19, 524–531 (2003).
Bernal, A., Ear, U. & Kyrpides, N. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 29, 126–127 (2001).
Alm, E. J. et al. The MicrobesOnline Web site for comparative genomics. Genome Res. 15, 1015–1022 (2005).
Rey, S. et al. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 33, D164–D168 (2005).
Wheeler, D. L. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36, D13–D21 (2008).
Overbeek, R. et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702 (2005).
Apweiler, R. et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 (2004).
Borodina, I., Krabben, P. & Nielsen, J. Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res. 15, 820–829 (2005).
Karp, P. D., Paley, S. & Romero, P. The Pathway Tools software. Bioinformatics 18, S225–S232 (2002).
Arakawa, K., Yamada, Y., Shinoda, K., Nakayama, Y. & Tomita, M. GEM System: automatic prototyping of cell-wide metabolic pathway models from genomes. BMC Bioinformatics 7, 168 (2006).
Pinney, J. W., Shirley, M. W., McConkey, G. A. & Westhead, D. R. metaSHARK: software for automated metabolic network prediction from DNA sequence and its application to the genomes of Plasmodium falciparum and Eimeria tenella. Nucleic Acids Res. 33, 1399–1409 (2005).
Goesmann, A., Haubrock, M., Meyer, F., Kalinowski, J. & Giegerich, R. PathFinder: reconstruction and dynamic visualization of metabolic pathways. Bioinformatics 18, 124–129 (2002).
Satish Kumar, V., Dasika, M. S. & Maranas, C. D. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics 8, 212 (2007).
Green, M. L. & Karp, P. D. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics 5, 76 (2004).
Henry, C. S., Jankowski, M. D., Broadbelt, L. J. & Hatzimanikatis, V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys. J. 90, 1453–1461 (2006).
Kümmel, A., Panke, S. & Heinemann, M. Systematic assignment of thermodynamic constraints in metabolic network models. BMC Bioinformatics 7, 512 (2006).
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008).
Pouliot, Y. & Karp, P. D. A survey of orphan enzyme activities. BMC Bioinformatics 8, 244 (2007).
Thomason, L. C., Court, D. L., Datta, A. R., Khanna, R. & Rosner, J. L. Identification of the Escherichia coli K-12 ybhE gene as pgl, encoding 6-phosphogluconolactonase. J. Bacteriol. 186, 8248–8253 (2004).
Fuhrer, T., Chen, L., Sauer, U. & Vitkup, D. Computational prediction and experimental verification of the gene encoding the NAD+/NADP+-dependent succinate semialdehyde dehydrogenase in Escherichia coli. J. Bacteriol. 189, 8073–8078 (2007).
Pinney, J. W. et al. Metabolic reconstruction and analysis for parasite genomes. Trends Parasitol. 23, 548–554 (2007). A detailed review of the challenges that are encountered in the reconstruction of metabolic networks for parasites such as P. falciparum.
Balu, B. & Adams, J. H. Advancements in transfection technologies for Plasmodium. Int. J. Parasitol. 37, 1–10 (2007).
Kirk, K. & Saliba, K. J. Targeting nutrient uptake mechanisms in Plasmodium. Curr. Drug Targets 8, 75–88 (2007).
Daily, J. P. et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 450, 1091–1095 (2007).
Deitsch, K. et al. Mechanisms of gene regulation in Plasmodium. Am. J. Trop. Med. Hyg. 77, 201–208 (2007).
Shlomi, T. et al. Systematic condition-dependent annotation of metabolic genes. Genome Res. 17, 1626–1633 (2007).
Saito, N. et al. Metabolomics approach for enzyme discovery. J. Proteome Res. 5, 1979–1987 (2006).
Chiang, K. P., Niessen, S., Saghatelian, A. & Cravatt, B. F. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem. Biol. 13, 1041–1050 (2006).
Popescu, L. & Yona, G. Automation of gene assignments to metabolic pathways using high-throughput expression data. BMC Bioinformatics 6, 217 (2005).
Rodionov, D. A. et al. Genomic identification and in vitro reconstitution of a complete biosynthetic pathway for the osmolyte di-myo-inositol-phosphate. Proc. Natl Acad. Sci. USA 104, 4279–4284 (2007).
Thiele, I., Jamshidi, N., Fleming, R. M. T. & Palsson, B. O. Genome-scale reconstruction of E. coli's transcriptional and translational machinery: a knowledge-base and its mathematical formulation. PLoS Comput. Biol. (in the press).
Acknowledgements
The authors thank A. Osterman and N. Jamshidi for their insights. A.M.F. and I.T. were supported by National Institutes of Health (NIH) grant R01 GM057089 and M.J.H. was supported by NIH grant R01 GM071808. B.O.P. serves on the scientific advisory board of Genomatica.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary information S1 (table)
Available predictive genome-scale metabolic network reconstructions (PDF 216 kb)
Supplementary information S2 (table)
Common issues encountered during metabolic network reconstructions (PDF 112 kb)
Related links
Glossary
- BiGG knowledge base
-
The collection of established biochemical, genetic and genomic data (BiGG) represented by a network reconstruction.
- Genome-scale network reconstruction
-
(GENRE). A two-dimensional genome annotation (for example, a metabolic reconstruction) that contains a list of all the chemical transformations known to take place in a particular network (usually the entire metabolic network of a particular organism; for example, a GENRE of E. coli). These transformations can be represented by a stoichiometric matrix. A genre is updated as the BiGG knowledge base expands.
- Genome-scale model
-
A network reconstruction in a mathematical format that can be computationally interrogated and can be subsequently used for experimental design.
- Bibliomic data
-
Legacy data that are contained in peer-reviewed scientific publications. The 'omic designation represents a comprehensive assessment of legacy data for a target organism.
- Stoichiometric matrix
-
A matrix that contains the stoichiometric coefficients for the reactions that constitute a network. The rows represent the compounds, the columns represent the chemical transformations and the entries represent the stoichiometric coefficients.
Rights and permissions
About this article
Cite this article
Feist, A., Herrgård, M., Thiele, I. et al. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol 7, 129–143 (2009). https://doi.org/10.1038/nrmicro1949
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1949
This article is cited by
-
Genome-scale metabolic network model and phenome of solvent-tolerant Pseudomonas putida S12
BMC Genomics (2024)
-
Systematizing Microbial Bioplastic Production for Developing Sustainable Bioeconomy: Metabolic Nexus Modeling, Economic and Environmental Technologies Assessment
Journal of Polymers and the Environment (2023)
-
Multi-genome metabolic modeling predicts functional inter-dependencies in the Arabidopsis root microbiome
Microbiome (2022)
-
Addressing uncertainty in genome-scale metabolic model reconstruction and analysis
Genome Biology (2021)
-
Transcriptome integrated metabolic modeling of carbon assimilation underlying storage root development in cassava
Scientific Reports (2021)