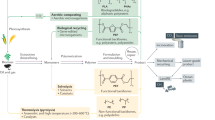
Key Points
-
Organophosphorus (OP) compounds are widely used as pesticides, petroleum additives and plasticizers. A large amount of chemical warfare agents (CWAs) also belong to the OP group. As a result of extensive and excessive use, environmental contamination with OP pesticides has been reported in several parts of the world.
-
Owing to high mammalian toxicity, OP waste and residues are excellent candidates for bioremediation. An effective therapy for intentional and unintentional OP poisoning is also needed.
-
Several bacteria, a few fungi and cyanobacteria with the capability to degrade OP compounds, using OP as a source of energy or co-metabolically, have been isolated from different corners of the globe. Some have been successfully used for bioremediation.
-
Most of the enzymes isolated from various phylogenetically different bacteria fall in three main types: OPH (OP hydrolase), MPH (methyl parathion hydrolase) and OP acid anhydrolase). These enzymes belong to three separate superfamilies.
-
The origin and evolution of OP-degrading enzymes are still a matter of debate. Recent findings suggest that at least OPH may have originated from the promiscuous activity of lactonase (an enzyme that hydrolyses lactones).
-
OP-degrading bacteria and their enzymes could form the basis of multiple biotechnological applications across several disciplines (such as bioremediation, and medical and genetic marker industries). However, to convert this promise into practice, emerging technologies, such as metagenomics and nanotechnology, together with conventional biochemical and molecular approaches, need to be adopted.
Abstract
The first organophosphorus (OP) compound-degrading bacterial strain was isolated from a paddy field in the Philippines in 1973. Since then, several phylogenetically distinct bacteria that can degrade OP by co-metabolism, or use OPs as a source of carbon, phosphorus or nitrogen, have been isolated from different parts of the world. There is huge potential for industrial applications of OP-degrading bacteria. Important advances in our understanding of the microbiology, genomics and evolution of OP-degrading bacteria have been made over the past four decades, and are discussed in this Review.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Dragun, J., Kuffner, A. C. & Schneiter, R. W. Groundwater contamination.1. Transport and transformations of organic chemicals. Chem. Engineer. 91, 65–70 (1984).
Organophosphate (Post note 12). Parliamentary Office of Science and Technology [online], (1998).
Ballantyne, B. & Marrs, T. C. Clinical and Experimental Toxicology of Organophosphates and Carbamates (Butterworth Heinemann, Oxford, 1992).
EPA. Review of chlorpyrifos poisoning data. US EPA 1–46 (1995).
Cisar, J. L. & Snyder, G. H. Fate and management of turfgrass chemicals. ACS Symp. Ser. 743, 106–126 (2000).
Boucard, T. K., Parry, J., Jones, K. & Semple, K. T. Effects of organophosphates and synthetic pyrethroid sheep dip formulations on protozoan survival and bacterial survival and growth. FEMS Microbiol. Ecol. 47, 121–127 (2004).
Galloway, T. & Handy, R. Immunotoxicity of organophosphorous pesticides. Ecotoxicology 12, 345–363 (2003).
Bird, S. et al. OpdA, a bacterial organophosphorus hydrolase, prevents lethality in rats after poisoning with highly toxic organophosphorus pesticides. Toxicology 247, 88–92 (2008).
Ragnarsdottir, K. V. Environmental fate and toxicology of organophosphate pesticides. J. Geol. Soc. London 157, 859–876 (2000).
Eddleston, M. et al. Management of acute organophosphorus pesticide poisoning. Lancet 371, 597–607 (2008). This article highlights the difficulties in medical management and inadequacies of current therapies for OP poisoning.
Lotti, M. Promotion of organophosphate induced delayed polyneuropathy by certain esterase inhibitors. Toxicology 181, 245–248 (2002).
Singh, B. K. & Walker, A. Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 30, 428–471 (2006). This article describes known and possible mechanisms of OP degradation by microorganisms in the environment.
Singh, B. K., Walker, A., Morgan, J. A. W. & Wright, D. J. Effects of soil pH on the biodegradation of chlorpyrifos and isolation of a chlorpyrifos-degrading bacterium. Appl. Environ. Microbiol. 69, 5198–5206 (2003).
Singh, B. K., Walker, A., Morgan, J. A. W. & Wright, D. J. Role of soil pH in the development of enhanced biodegradation of fenamiphos. Appl. Environ. Microbiol. 69, 7035–7043 (2003).
Price, O. R., Walker, A., Wood, M. & Oliver, M. A. in Proc. XII Symp. Pest. Chem. 73–82 (La Goliardica Pavese, Pavia, 2003).
Singh, B. K., Walker, A. & Wright, D. J. Cross-enhancement of accelerated biodegradation of organophosphorus compounds in soils: dependence on structural similarity of compounds. Soil Biol. Biochem. 37, 1675–1682 (2005).
Watanabe, K. & Hamamura, N. Molecular and physiological approaches to understanding the ecology of pollutant degradation. Curr. Opin. Biotechnol. 14, 289–295 (2003).
Sethunathan, N. & Yoshida, T. Flavobacterium sp. that degrades diazinon and parathion. Can. J. Microbiol. 19, 873–875 (1973). This is the first report of the isolation of OP-degrading bacteria from the environment. A number of other microorganisms with similar degrading capabilities have since been isolated.
Serdar, C. M., Gibson, D. T., Munnecke, D. M. & Lancaster, J. H. Plasmid involvement in parathion hydrolysis by Pseudomonas diminuta. Appl. Environ. Microbiol. 44, 246–249 (1982).
Siddavattam, D., Khajamohiddin, S., Manavathi, B., Pakala, S. B. & Merrick, M. Transposon-like organization of the plasmid-borne organophosphate degradation (opd) gene cluster found in Flavobacterium sp. Appl. Environ. Microbiol. 69, 2533–2539 (2003). This study describes the genetic structure of the opd operon in Flavobacterium sp. ATCC 27551, which was shown to contain a transposon gene.
Horne, I., Sutherland, T. D., Harcourt, R. L., Russell, R. J. & Oakeshott, J. G. Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate. Appl. Environ. Microbiol. 68, 3371–3376 (2002).
Mulbry, W. W. & Karns, J. S. Parathion hydrolase specified by the Flavobacterium opd gene: relationship between the gene and protein. J. Bacteriol. 171, 6740–6746 (1989).
Raushel, F. M. Bacterial detoxification of organophosphate nerve agents. Curr. Opin. Microbiol. 5, 288–295 (2002). This review describes the origin and mode of action of OPH enzymes and highlights possible mechanisms to improve the efficacy of the enzymes against poor substrates.
Horne, I., Qiu, X. H., Russell, R. J. & Oakeshott, J. G. The phosphotriesterase gene opdA in Agrobacterium radiobacter p230 is transposable. FEMS Microbiol. Lett. 222, 1–8 (2003).
Yang, H. et al. Evolution of an organophosphate-degrading enzyme: a comparison of natural and directed evolution. Protein Eng. 16, 135–145 (2003).
Gressel, J. & Levy, A. A. Agriculture: the selector of improbable mutations. Proc. Natl Acad. Sci. USA 103, 12215–12216 (2006).
Lynch, M. Simple evolutionary pathways to complex proteins. Protein Sci. 14, 2217–2225 (2005).
Afriat, L., Roodveldt, C., Manco, G. & Tawfik, D. S. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 45, 13677–13686 (2006). This study postulates that phosphotriesterase evolved from lactonase and provides experimental evidence to support this hypothesis.
Elias, M. et al. Crystallization and preliminary X-ray diffraction analysis of the hyperthermophilic Sulfolobus solfataricus phosphotriesterase. Acta Crystallogr. 63, 553–555 (2007).
Elias, M. et al. Structural basis for natural lactonase and promiscuous phosphotriesterase activities. J. Mol. Biol. 379, 1017–1028 (2008).
Aharoni, A. et al. The 'evolvability' of promiscuous protein functions. Nature Genet. 37, 73–76 (2005).
Chen, L. M. et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 187, 4992–4999 (2005).
Porzio, E., Merone, L., Mandrich, L., Rossi, M. & Manco, G. A new phosphotriesterase from Sulfolobus acidocaldarius and its comparison with the homologue from Sulfolobus solfataricus. Biochimie 89, 625–636 (2007).
Dong, Y. J. et al. Crystal structure of methyl parathion hydrolase from Pseudomonas sp. WBC-3. J. Mol. Biol. 353, 655–663 (2005).
Cheng, T.-C., Harvey, S. P. & Stroup, A. N. Purification and properties of a highly active organophosphorus acid anhydrolase from Alteromonas undina. Appl. Environ. Microbiol. 59, 3138–3140 (1993).
Cheng, T.-C., DeFrank, J. J. & Rastogi, V. K. Alteromonas prolidase for organophosphorus G-agent decontamination. Chem. Biol. Interact. 120, 455–462 (1999).
Mulbry, W. W., Kearney, P. C., Nelson, J. O. & Karns, J. S. Physical comparison of parathion hydrolase plasmids from Pseudomonas diminuta and Flavobacterium sp. Plasmid 18, 173–177 (1987).
Zhang, R. F. et al. Cloning of the organophosphorus pesticide hydrolase gene clusters of seven degradative bacteria isolated from a methyl parathion contaminated site and evidence of their horizontal gene transfer. Biodegradation 17, 465–472 (2006).
Liebert, C. A., Hall, R. M. & Summers, A. O. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63, 507–522 (1999).
Frost, L. S., Leplae, R., Summers, A. O. & Toussaint, A. Mobile genetic elements: the agents of open source evolution. Nature Rev. Microbiol. 3, 722–732 (2005).
Mulbry, W., Ahrens, E. & Karns, J. Use of a field-scale biofilter for the degradation of the organophosphate insecticide coumaphos in cattle dip wastes. Pestic. Sci. 52, 268–274 (1998). This is the first report of the successful use of bioremediation for the removal of OP waste on a large scale.
Karns, J. S., Hapeman, C. J., Mulbry, W. W., Ahrens, E. H. & Shelton, D. R. Biotechnology for the elimination of agrochemical wastes. HortScience 33, 626–631 (1998).
Walker, A. W. & Keasling, J. D. Metabolic engineering of Pseudomonas putida for the utilization of parathion as a carbon and energy source. Biotechnol. Bioeng. 78, 715–721 (2002).
Zheng, Y. Z., Lan, W. S., Qiao, C. L., Mulchandani, A. & Chen, W. Decontamination of vegetables sprayed with organophosphate pesticides by organophosphorus hydrolase and carboxylesterase (B1). Appl. Biochem. Biotechnol. 136, 233–241 (2007).
Wang, X. X. et al. Phytodegradation of organophosphorus compounds by transgenic plants expressing a bacterial organophosphorus hydrolase. Biochem. Biophys. Res. Commun. 365, 453–458 (2008).
Lee, J. H. et al. Stable and continuous long-term enzymatic reaction using an enzyme–nanofiber composite. Appl. Microbiol. Biotechnol. 75, 1301–1307 (2007).
Kim, J. & Grate, J. W. Single-enzyme nanoparticles armored by a nanometer-scale organic/inorganic network. Nano Lett. 3, 1219–1222 (2003).
Jia, H. F., Zhu, G. Y. & Wang, P. Catalytic behaviors of enzymes attached to nanoparticles: the effect of particle mobility. Biotechnol. Bioeng. 84, 406–414 (2003).
Mulchandani, A., Mulchandani, P., Kanifar, H. & Chen, W. Direct monitoring of organophosphorus nerve agents by amperometric enzyme biosensor. Abstr. Amer. Chem. Soc. 217, U789–U790 (1999).
Wang, J., Chen, L., Mulchandani, A., Mulchandani, P. & Chen, W. Remote biosensor for in-situ monitoring of organophosphate nerve agents. Electroanalysis 11, 866–869 (1999).
Wang, J. et al. Dual amperometric–potentiometric biosensor detection system for monitoring organophosphorus neurotoxins. Anal. Chim. Acta 469, 197–203 (2002).
Liu, N. Y. et al. Single-walled carbon nanotube based real-time organophosphate detector. Electroanalysis 19, 616–619 (2007).
Yang, W. et al. Application of methyl parathion hydrolase (MPH) as a labeling enzyme. Anal. Bioanal. Chem. 390, 2133–2140 (2008).
Pinkerton, T. S., Howard, J. A. & Wild, J. R. Genetically engineered resistance to organophosphate herbicides provides a new scoreable and selectable marker system for transgenic plants. Mol. Breed. 21, 27–36 (2008).
Sogorb, M. A., Vilanova, E. & Carrera, V. Future applications of phosphotriesterases in the prophylaxis and treatment of organophosporus insecticide and nerve agent poisonings. Toxicol. Lett. 151, 219–233 (2004).
Sogorb, M. A. & Vilanova, E. Enzymes involved in the detoxification of organophosphorus, carbamate and pyrethroid insecticides through hydrolysis. Toxicol. Lett. 128, 215–228 (2002).
Eyer, P. The role of oximes in management of organophosphorus pesticide poisoning. Toxicol. Rev. 22, 165–190 (2003).
Petrikovics, I. et al. Antagonism of paraoxon intoxication by recombinant phosphotriesterase encapsulated within sterically stabilized liposomes. Toxicol. Appl. Pharmacol. 156, 56–63 (1999).
Mann, J. F. S. et al. Optimisation of a lipid based oral delivery system containing A/Panama influenza haemagglutinin. Vaccine 22, 2425–2429 (2004).
Hirsch, L. R. et al. Nanoshell-mediated near-infrared thermal therapy of tumors under magnetic resonance guidance. Proc. Natl Acad. Sci. USA 100, 13549–13554 (2003).
Schloss, P. D. & Handelsman, J. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14, 303–310 (2003). This article provides a good overview of the capability of metagenomics to isolate industrial enzymes from unculturable bacteria.
Singh, B. K., Millard, P., Whiteley, A. S. & Murrell, J. C. Unravelling rhizosphere–microbial interactions: opportunities and limitations. Trends Microbiol. 12, 386–393 (2004).
Lee, N. et al. Combination of fluorescent in situ hybridization and microautoradiography — a new tool for structure–function analyses in microbial ecology. Appl. Environ. Microbiol. 65, 1289–1297 (1999).
Gray, N. D. & Head, I. M. Linking genetic identity and function in communities of uncultured bacteria. Environ. Microbiol. 3, 481–492 (2001).
Radajewski, S., Ineson, P., Parekh, N. R. & Murrell, J. C. Stable-isotope probing as a tool in microbial ecology. Nature 403, 646–649 (2000).
Neufeld, J. D., Wagner, M. & Murrell, J. C. Who eats what, where and when? Isotope-labelling experiments are coming of age. ISME J. 1, 103–110 (2007).
Manefield, M. & Whiteley, A. S. Acylated homoserine lactones in the environment: chameleons of bioactivity. Philos. Trans. R. Soc. Lond. B 362, 1235–1240 (2007).
Roche, D. et al. Communications blackout? Do N-acylhomoserine-lactone-degrading enzymes have any role in quorum sensing? Microbiology 150, 2023–2028 (2004).
Acknowledgements
I thank A. Taylor (Macaulay Institute), G. Elliott (Macaulay Institute), I. Anderson (University of Western Sydney) and G. Bending (Warwick HRI) for their comments on the manuscript, and P. Millard (Macaulay Institute) and C. Macdonald (Rothamsted Research) for detailed discussions. Work in my laboratory is funded by the Scottish Government.
Author information
Authors and Affiliations
Related links
Glossary
- Bioremediation
-
A biological process that uses living organisms or their products (enzymes) to convert a harmful substance to a non-toxic substance or to return the contaminated environment to its original condition.
- Biosensor
-
A detector device made from a biological component combined with a physico–chemical detector that is used for detection of a substance or chemical.
- Biodegradation
-
A process by which an indigenous bacterial population acquires genes that encode enzymes to allow the use of xenobiotics as an energy source.
- Xenobiotic
-
A chemical that is usually man-made and is not found naturally in the environment.
- Bioreactor
-
A device or system that supports a biologically active environment.
- Nanoparticle
-
A small particle (one or more dimensions of ∼100 nM or less) that behaves as a whole unit; for example, in terms of transportation.
- Quantum dot
-
A type of nanoparticle that can be used for optical, electrical, biological and medical purposes.
Rights and permissions
About this article
Cite this article
Singh, B. Organophosphorus-degrading bacteria: ecology and industrial applications. Nat Rev Microbiol 7, 156–164 (2009). https://doi.org/10.1038/nrmicro2050
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2050
This article is cited by
-
Potential of nano-phytoremediation of heavy metal contaminated soil: emphasizing the role of mycorrhizal fungi in the amelioration process
International Journal of Environmental Science and Technology (2024)
-
Biodegradation of monocrotophos by Brucella intermedia Msd2 isolated from cotton plant
World Journal of Microbiology and Biotechnology (2023)
-
Data-informed discovery of hydrolytic nanozymes
Nature Communications (2022)
-
Evidence for enhanced dissipation of chlorpyrifos in an agricultural soil inoculated with Serratia rubidaea strain ABS 10
Environmental Science and Pollution Research (2022)
-
Structural design of nanosize-metal–organic framework-based sensors for detection of organophosphorus pesticides in food and water samples: current challenges and future prospects
Journal of Nanostructure in Chemistry (2022)