Key Points
-
The Sec machinery is essential for life. All cells need to assemble phospholipid bilayer membranes, which have embedded proteins. In bacteria, the Sec pathway catalyses most of the load of protein secretion and acts as the front end for several subsequent protein-sorting and sub-cellular-targeting machines.
-
A combination of membrane-embedded and soluble factors that contribute to pre-protein targeting and translocation are described. A membrane-embedded pre-protein-conducting channel and an ATPase motor lie at its core.
-
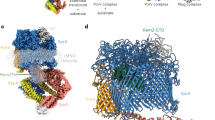
Atomic resolution structures of the pre-protein-conducting channel, its ATPase motor and targeting chaperones are available.
-
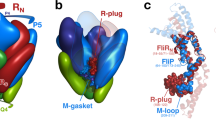
The protein-conducting channel is composed of several tilted and straight helices of varying lengths and is gated by a periplasmic plug. It has a well-characterized closed state and an anticipated open state that is expected to result from dilation.
-
Metabolic energy in the form of both ATP and the proton motive force is used to power pre-protein movement through the translocase machine.
-
The available data allow for a synthesis of multiple sub-reactions into a coherent model. This model describes how the translocase recognizes secretory proteins at specific sites and how it subsequently promotes protein export by a series of distinct energy-driven conformational states.
Abstract
All cells must traffic proteins across their membranes. This essential process is responsible for the biogenesis of membranes and cell walls, motility and nutrient scavenging and uptake, and is also involved in pathogenesis and symbiosis. The translocase is an impressively dynamic nanomachine that is the central component which catalyses transmembrane crossing. This complex, multi-stage reaction involves a cascade of inter- and intramolecular interactions that select, sort and target polypeptides to the membrane, and use energy to promote the movement of these polypeptides across — or their lateral escape and integration into — the phospholipid bilayer, with high fidelity and efficiency. Here, we review the most recent data on the structure and function of the translocase nanomachine.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Economou, A. et al. Secretion by numbers: protein traffic in prokaryotes. Mol. Microbiol. 62, 308–319 (2006).
Holland, I. B. Translocation of bacterial proteins — an overview. Biochim. Biophys. Acta 1694, 5–16 (2004).
Luirink, J. & Sinning, I. SRP-mediated protein targeting: structure and function revisited. Biochim. Biophys. Acta 1694, 17–35 (2004).
Randall, L. L. & Hardy, S. J. SecB, one small chaperone in the complex milieu of the cell. Cell. Mol. Life Sci. 59, 1617–1623 (2002).
Van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004). The first crystal structure of the membrane-embedded pre-protein-conducting channel. It provided hypotheses for the channel-dilation mechanism and its gating.
Li, W. et al. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol. Cell 26, 511–521 (2007).
Hanein, D. et al. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell 87, 721–732 (1996).
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001).
Beckmann, R. et al. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science 278, 2123–2126 (1997).
Manting, E. H., van Der Does, C., Remigy, H., Engel, A. & Driessen, A. J. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 19, 852–861 (2000).
Scheuring, J. et al. The oligomeric distribution of SecYEG is altered by SecA and translocation ligands. J. Mol. Biol. 354, 258–271 (2005).
Breyton, C., Haase, W., Rapoport, T. A., Kuhlbrandt, W. & Collinson, I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665 (2002).
Mitra, K. et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 (2005).
Hunt, J. F. et al. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297, 2018–2026 (2002). The first crystal structure of the SecA motor subunit of the translocase. Provided high-resolution information for the structural and functional dissection of the motor.
Osborne, A. R., Clemons, W. M. Jr & Rapoport, T. A. A large conformational change of the translocation ATPase SecA. Proc. Natl Acad. Sci. USA 101, 10937–10942 (2004).
Zimmer, J., Li, W. & Rapoport, T. A. A novel dimer interface and conformational changes revealed by an X-ray structure of B. subtilis SecA. J. Mol. Biol. 364, 259–265 (2006).
Papanikolau, Y. et al. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J. Mol. Biol. 366, 1545–1557 (2007).
Sharma, V. et al. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc. Natl Acad. Sci. USA 100, 2243–2248 (2003).
Vassylyev, D. G. et al. Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J. Mol. Biol. 364, 248–258 (2006).
Ullers, R. S. et al. SecB is a bona fide generalized chaperone in Escherichia coli. Proc. Natl Acad. Sci. USA 101, 7583–7588 (2004).
Schierle, C. F. et al. The DsbA signal sequence directs efficient, cotranslational export of passenger proteins to the Escherichia coli periplasm via the signal recognition particle pathway. J. Bacteriol. 185, 5706–5713 (2003).
Sijbrandi, R. et al. Signal recognition particle (SRP)-mediated targeting and Sec-dependent translocation of an extracellular Escherichia coli protein. J. Biol. Chem. 278, 4654–4659 (2003).
Randall, L. L. Translocation of domains of nascent periplasmic proteins across the cytoplasmic membrane is independent of elongation. Cell 33, 231–240 (1983).
Hartl, F. U., Lecker, S., Schiebel, E., Hendrick, J. P. & Wickner, W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63, 269–279 (1990).
Scotti, P. A. et al. SecA is not required for signal recognition particle-mediated targeting and initial membrane insertion of a nascent inner membrane protein. J. Biol. Chem. 274, 29883–29888 (1999).
Neumann-Haefelin, C., Schafer, U., Muller, M. & Koch, H. G. SRP-dependent co-translational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 19, 6419–6426 (2000).
Paetzel, M., Karla, A., Strynadka, N. C. & Dalbey, R. E. Signal peptidases. Chem. Rev. 102, 4549–4580 (2002).
Mogensen, J. E. & Otzen, D. E. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 57, 326–346 (2005).
Nakamoto, H. & Bardwell, J. C. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim. Biophys. Acta 1694, 111–119 (2004).
Gruber, C. W., Cemazar, M., Heras, B., Martin, J. L. & Craik, D. J. Protein disulfide isomerase: the structure of oxidative folding. Trends Biochem. Sci. 31, 455–464 (2006).
Brundage, L., Hendrick, J. P., Schiebel, E., Driessen, A. J. & Wickner, W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62, 649–657 (1990). In vitro functional reconstitution of the translocase from purified components. This study identified the components of the core channel and opened the floodgates for subsequent biochemical and biophysical studies.
Bostina, M., Mohsin, B., Kuhlbrandt, W. & Collinson, I. Atomic model of the E. coli membrane-bound protein translocation complex SecYEG. J. Mol. Biol. 352, 1035–1043 (2005).
Schatz, P. J., Bieker, K. L., Ottemann, K. M., Silhavy, T. J. & Beckwith, J. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 10, 1749–1757 (1991).
Saparov, S. M. et al. Determining the conductance of the SecY protein translocation channel for small molecules. Mol. Cell 26, 501–509 (2007).
Cannon, K. S., Or, E., Clemons, W. M. Jr, Shibata, Y. & Rapoport, T. A. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 169, 219–225 (2005).
Bieker, K. L., Phillips, G. J. & Silhavy, T. J. The sec and prl genes of Escherichia coli. J. Bioenerg. Biomembr. 22, 291–310 (1990).
Derman, A. I., Puziss, J. W., Bassford, P. J. Jr & Beckwith, J. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J. 12, 879–888 (1993).
Plath, K., Mothes, W., Wilkinson, B. M., Stirling, C. J. & Rapoport, T. A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94, 795–807 (1998).
Tam, P. C., Maillard, A. P., Chan, K. K. & Duong, F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 24, 3380–3388 (2005).
Maillard, A. P., Lalani, S., Silva, F., Belin, D. & Duong, F. Deregulation of the SecYEG translocation channel upon removal of the plug domain. J. Biol. Chem. 282, 1281–1287 (2007).
Junne, T., Schwede, T., Goder, V. & Spiess, M. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol. Biol. Cell 17, 4063–4068 (2006).
Tani, K., Tokuda, H. & Mizushima, S. Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli. Effect of membrane energization. J. Biol. Chem. 265, 17341–17347 (1990).
De Keyzer, J., Van Der Does, C. & Driessen, A. J. Kinetic analysis of the translocation of fluorescent precursor proteins into Escherichia coli membrane vesicles. J. Biol. Chem. 277, 46059–46065 (2002).
Gumbart, J. & Schulten, K. Molecular dynamics studies of the archaeal translocon. Biophys. J. 90, 2356–2367 (2006).
Nishiyama, K., Suzuki, T. & Tokuda, H. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell 85, 71–81 (1996).
Alami, M., Dalal, K., Lelj-Garolla, B., Sligar, S. G. & Duong, F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 26, 1995–2004 (2007).
Mori, H. et al. Fluorescence resonance energy transfer analysis of protein translocase. SecYE from Thermus thermophilus HB8 forms a constitutive oligomer in membranes. J. Biol. Chem. 278, 14257–14264 (2003).
Hamman, B. D., Chen, J. C., Johnson, E. E. & Johnson, A. E. The aqueous pore through the translocon has a diameter of 40–60 Å during cotranslational protein translocation at the ER membrane. Cell 89, 535–544 (1997).
Osborne, A. R. & Rapoport, T. A. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell 129, 97–110 (2007). Describes how in dimeric SecYEG one trimer provides the active exit pore and SecA docks on the other trimer.
Baud, C. et al. Allosteric communication between signal peptides and the SecA protein DEAD motor ATPase domain. J. Biol. Chem. 277, 13724–13731 (2002).
Karamanou, S. et al. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol. Microbiol. 34, 1133–1145 (1999).
Papanikou, E. et al. Identification of the preprotein binding domain of SecA. J. Biol. Chem. 280, 43209–43217 (2005).
Sianidis, G. et al. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 20, 961–970 (2001).
Price, A., Economou, A., Duong, F. & Wickner, W. Separable ATPase and membrane insertion domains of the SecA subunit of preprotein translocase. J. Biol. Chem. 271, 31580–31584 (1996).
Keramisanou, D. et al. Disorder-order folding transitions underlie catalysis in the helicase motor of SecA. Nature Struct. Mol. Biol. 13, 594–602 (2006).
Cordin, O., Banroques, J., Tanner, N. K. & Linder, P. The DEAD-box protein family of RNA helicases. Gene 367, 17–37 (2006).
Mori, H. & Ito, K. The long alpha-helix of SecA is important for the ATPase coupling of translocation. J. Biol. Chem. 281, 36249–36256 (2006).
Breukink, E. et al. The C terminus of SecA is involved in both lipid binding and SecB binding. J. Biol. Chem. 270, 7902–7907 (1995).
Matousek, W. M. & Alexandrescu, A. T. NMR structure of the C-terminal domain of SecA in the free state. Biochim. Biophys. Acta 1702, 163–171 (2004).
Dempsey, B. R. et al. Solution NMR structure and X-ray absorption analysis of the C-terminal zinc-binding domain of the SecA ATPase. Biochemistry 43, 9361–9371 (2004).
Fekkes, P., de Wit, J. G., Boorsma, A., Friesen, R. H. & Driessen, A. J. Zinc stabilizes the SecB binding site of SecA. Biochemistry 38, 5111–5116 (1999).
Musial-Siwek, M., Rusch, S. L. & Kendall, D. A. Selective photoaffinity labeling identifies the signal peptide binding domain on SecA. J. Mol. Biol. 365, 637–648 (2007).
Gelis, I. et al. Structural basis for signal sequence recognition by the 204kDa translocase motor SecA determined by NMR. Cell (in the press). This study presents the first structure of a pre-protein segment with a translocase component and reveals movements in the PBD that could underlie the mechanism of translocation. This is the first solution structure of SecA. Owing to its size, this was a major challenge for NMR-based methods.
Karamanou, S. et al. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 26, 2904–2914 (2007). First insight into how pre-proteins control ATP-driven catalysis by the SecA motor through the PBD.
Vrontou, E., Karamanou, S., Baud, C., Sianidis, G. & Economou, A. Global co-ordination of protein translocation by the SecA IRA1 switch. J. Biol. Chem. 279, 22490–22497 (2004).
Woodbury, R. L., Hardy, S. J. & Randall, L. L. Complex behavior in solution of homodimeric SecA. Protein Sci. 11, 875–882 (2002).
Driessen, A. J. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry 32, 13190–13197 (1993).
Shilton, B. et al. Escherichia coli SecA shape and dimensions. FEBS Lett. 436, 277–282 (1998).
Shinkai, A., Akita, M., Matsuyama, S. & Mizushima, S. Quantitative renaturation from a guanidine-denatured state of the SecA dimer, a 200 KDa protein involved in protein secretion in Escherichia coli. Biochem. Biophys. Res. Commun. 172, 1217–1223 (1990).
Akita, M., Shinkai, A., Matsuyama, S. & Mizushima, S. SecA, an essential component of the secretory machinery of Escherichia coli, exists as homodimer. Biochem. Biophys. Res. Commun. 174, 211–216 (1991).
Dempsey, B. R., Economou, A., Dunn, S. D. & Shilton, B. H. The ATPase domain of SecA can form a tetramer in solution. J. Mol. Biol. 315, 831–843 (2002).
Hirano, M., Matsuyama, S. & Tokuda, H. The carboxyl-terminal region is essential for Sec-A dimerization. Biochem. Biophys. Res. Commun. 229, 90–95 (1996).
Karamanou, S. et al. Escherichia coli SecA truncated at its termini is functional and dimeric. FEBS Lett. 579, 1267–1271 (2005).
Mori, H. et al. Amino-terminal region of SecA is involved in the function of SecG for protein translocation into Escherichia coli membrane vesicles. J. Biochem. (Tokyo) 124, 122–129 (1998).
Or, E., Boyd, D., Gon, S., Beckwith, J. & Rapoport, T. The bacterial ATPase SecA functions as a monomer in protein translocation. J. Biol. Chem. 280, 9097–9105 (2005).
Mori, H. & Ito, K. Different modes of SecY–SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc. Natl Acad. Sci. USA 103, 16159–16164 (2006).
van der Sluis, E. O. et al. Identification of two interaction sites in SecY that are important for the functional interaction with SecA. J. Mol. Biol. 361, 839–849 (2006).
Nagamori, S., Nishiyama, K. & Tokuda, H. Membrane topology inversion of SecG detected by labeling with a membrane-impermeable sulfhydryl reagent that causes a close association of SecG with SecA. J. Biochem. (Tokyo) 132, 629–634 (2002).
Nishiyama, K., Mizushima, S. & Tokuda, H. Preferential interaction of Sec-G with Sec-E stabilizes an unstable Sec-E derivative in the Escherichia coli cytoplasmic membrane. Biochem. Biophys. Res. Commun. 217, 217–223 (1995).
Snyders, S., Ramamurthy, V. & Oliver, D. Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J. Biol. Chem. 272, 11302–11306 (1997).
Dapic, V. & Oliver, D. Distinct membrane binding properties of N- and C-terminal domains of Escherichia coli SecA ATPase. J. Biol. Chem. 275, 25000–25007 (2000).
de Keyzer, J. et al. Covalently dimerized SecA is functional in protein translocation. J. Biol. Chem. 280, 35255–35260 (2005).
Jilaveanu, L. B. & Oliver, D. SecA dimer cross-linked at its subunit interface is functional for protein translocation. J. Bacteriol. 188, 335–338 (2006).
Or, E., Navon, A. & Rapoport, T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 21, 4470–4479 (2002).
Duong, F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 22, 4375–4384 (2003).
Benach, J. et al. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J. Biol. Chem. 278, 3628–3638 (2003).
Lill, R., Dowhan, W. & Wickner, W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 60, 271–280 (1990).
Tziatzios, C. et al. The bacterial protein-translocation complex: SecYEG dimers associate with one or two SecA molecules. J. Mol. Biol. 340, 513–524 (2004).
Collinson, I. et al. Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J. 20, 2462–2471 (2001).
Duong, F. & Wickner, W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 16, 2756–2768 (1997).
Pogliano, J. A. & Beckwith, J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 13, 554–561 (1994).
Matsuyama, S., Fujita, Y. & Mizushima, S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 12, 265–270 (1993).
Economou, A., Pogliano, J. A., Beckwith, J., Oliver, D. B. & Wickner, W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83, 1171–1181 (1995).
Yi, L. & Dalbey, R. E. Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria. Mol. Membr. Biol. 22, 101–111 (2005).
Serek, J. et al. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 23, 294–301 (2004).
Beck, K. et al. YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2, 709–714 (2001).
Nouwen, N. & Driessen, A. J. SecDFyajC forms a heterotetrameric complex with YidC. Mol. Microbiol. 44, 1397–1405 (2002).
van Dalen, A. & de Kruijff, B. The role of lipids in membrane insertion and translocation of bacterial proteins. Biochim. Biophys. Acta 1694, 97–109 (2004).
Suzuki, H., Nishiyama, K. & Tokuda, H. Increases in acidic phospholipid contents specifically restore protein translocation in a cold-sensitive secA or secG null mutant. J. Biol. Chem. 274, 31020–31024 (1999).
Rietveld, A. G., Killian, J. A., Dowhan, W. & de Kruijff, B. Polymorphic regulation of membrane phospholipid composition in Escherichia coli. J. Biol. Chem. 268, 12427–12433 (1993).
Xu, Z. et al. Crystal structure of the bacterial protein export chaperone SecB. Nature Struct. Biol. 12, 1172–1177 (2000).
Zhou, J. & Xu, Z. Structural determinants of SecB recognition by SecA in bacterial protein translocation. Nature Struct. Biol. 10, 942–947 (2003).
Dekker, C., de Kruijff, B. & Gros, P. Crystal structure of SecB from Escherichia coli. J. Struct. Biol. 144, 313–319 (2003).
Randall, L. L. et al. The interaction between the chaperone SecB and its ligands: evidence for multiple subsites for binding. Protein Sci. 7, 2384–2390 (1998).
Knoblauch, N. T. et al. Substrate specificity of the SecB chaperone. J. Biol. Chem. 274, 34219–34225 (1999).
Woodbury, R. L. et al. Complexes between protein export chaperone SecB and SecA. Evidence for separate sites on SecA providing binding energy and regulatory interactions. J. Biol. Chem. 275, 24191–24198 (2000).
Fekkes, P., van der Does, C. & Driessen, A. J. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 16, 6105–6113 (1997).
Crane, J. M. et al. Mapping of the docking of SecA onto the chaperone SecB by site-directed spin labeling: insight into the mechanism of ligand transfer during protein export. J. Mol. Biol. 353, 295–307 (2005).
Karamyshev, A. L. & Johnson, A. E. Selective SecA association with signal sequences in ribosome-bound nascent chains: a potential role for SecA in ribosome targeting to the bacterial membrane. J. Biol. Chem. 280, 37930–37940 (2005).
Eisner, G., Koch, H. G., Beck, K., Brunner, J. & Muller, M. Ligand crowding at a nascent signal sequence. J. Cell Biol. 163, 35–44 (2003).
Chou, Y. T. & Gierasch, L. M. The conformation of a signal peptide bound by Escherichia coli preprotein translocase SecA. J. Biol. Chem. 280, 32753–32760 (2005).
Khatib, K. & Belin, D. A novel class of secA alleles that exert a signal-sequence-dependent effect on protein export in Escherichia coli. Genetics 162, 1031–1043 (2002).
Kourtz, L. & Oliver, D. Tyr-326 plays a critical role in controlling SecA-preprotein interaction. Mol. Microbiol. 37, 1342–1356 (2000).
Baud, C. et al. Purification of a functional mature region from a SecA-dependent preprotein. Protein Expr. Purif. 40, 336–339 (2005).
Schiebel, E., Driessen, A. J., Hartl, F. U. & Wickner, W. ΔμH+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64, 927–939 (1991). Biochemical dissection of the distinct energetic roles of ATP and the PMF in promoting translocation.
Fak, J. J. et al. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry 43, 7307–7327 (2004).
Economou, A. & Wickner, W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78, 835–843 (1994). Demonstrated that ATP energy is used for defined SecA conformational events that correlate with the defined movements of short pre-protein segments described by Schiebel and colleagues in reference 115.
Yamada, H., Matsuyama, S., Tokuda, H. & Mizushima, S. A high concentration of SecA allows proton motive force-independent translocation of a model secretory protein into Escherichia coli membrane vesicles. J. Biol. Chem. 264, 18577–18581 (1989).
Nishiyama, K., Fukuda, A., Morita, K. & Tokuda, H. Membrane deinsertion of SecA underlying proton motive force-dependent stimulation of protein translocation. EMBO J. 18, 1049–1058 (1999).
Bessonneau, P., Besson, V., Collinson, I. & Duong, F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 21, 995–1003 (2002).
Nouwen, N., de Kruijff, B. & Tommassen, J. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc. Natl Acad. Sci. USA 93, 5953–5957 (1996).
Arkowitz, R. A., Joly, J. C. & Wickner, W. Translocation can drive the unfolding of a preprotein domain. EMBO J. 12, 243–253 (1993).
Engelman, D. M. & Steitz, T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell 23, 411–422 (1981).
Joly, J. C. & Wickner, W. The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids. EMBO J. 12, 255–263 (1993). First direct demonstration that pre-proteins travel in a proteinaceous environment that is composed of SecA and SecY and form distinct interactions with each.
Economou, T. & Dalbey, R. E. (eds) Special issue of Biochimica et Biophysica Acta — Molecular Cell Research on Protein Export/Secretion in Bacteria Vol. 1694, 1–333 (2004).
White, S. H. & von Heijne, G. Transmembrane helices before, during, and after insertion. Curr. Opin. Struct. Biol. 15, 378–386 (2005).
Kiefer, D. & Kuhn, A. Hydrophobic forces drive spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J. 18, 6299–6306 (1999).
Gallusser, A. & Kuhn, A. Initial steps in protein membrane insertion. Bacteriophage M13 procoat protein binds to the membrane surface by electrostatic interaction. EMBO J. 9, 2723–2729 (1990).
Urbanus, M. L. et al. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2, 524–529 (2001).
van der Laan, M., Bechtluft, P., Kol, S., Nouwen, N. & Driessen, A. J. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J. Cell Biol. 165, 213–222 (2004).
Clantin, B. et al. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317, 957–961 (2007).
Kim, S. et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 (2007).
Bos, M. P., Robert, V. & Tommassen, J. Biogenesis of the Gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 7 June 2007 (doi:10.1146/annurev.micro.61.080706.093245).
Crane, J. M. et al. Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling. J. Mol. Biol. 363, 63–74 (2006).
Acknowledgements
We are grateful to the Kalodimos laboratory for an exciting collaboration and to B. Kalodimos and T. Pugsley for stimulating discussions. Research in our laboratory is supported by grants from the European Union (LSHG-CT-2005-037586), the Greek General Secretariat of Research and the European Regional Development Fund (01AKMON46 and PENED03ED623).
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Entrez Genome Project
Entrez Protein
Protein Data Bank
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Papanikou, E., Karamanou, S. & Economou, A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol 5, 839–851 (2007). https://doi.org/10.1038/nrmicro1771
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1771
This article is cited by
-
The genome of Candidatus phytoplasma ziziphi provides insights into their biological characteristics
BMC Plant Biology (2023)
-
Arabinan saccharification by biogas reactor metagenome-derived arabinosyl hydrolases
Biotechnology for Biofuels and Bioproducts (2022)
-
Kaolinite weakens the co-stress of ampicillin and tetracycline on Escherichia coli through multiple pathways
Environmental Science and Pollution Research (2021)
-
In Silico Study of Different Signal Peptides to Express Recombinant Glutamate Decarboxylase in the Outer Membrane of Escherichia coli
International Journal of Peptide Research and Therapeutics (2020)
-
Genome-Wide Analysis of Putative G-Quadruplex Sequences (PGQSs) in Onion Yellows Phytoplasma (Strain OY-M): An Emerging Plant Pathogenic Bacteria
Indian Journal of Microbiology (2019)