Key Points
-
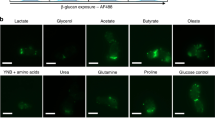
G-protein-coupled receptors (GPCRs) function as both pheromone and glucose sensors in fungi, whereas transceptors such as Snf3 and Rgt2 are involved in glucose sensing. Fungi have evolved sophisticated amino-acid-sensing systems including Ssy1–Ptr3–Ssy5 (SPS), Gap1 and GPCRs. The transceptors Pho84 and Pho87 have key roles in phosphate sensing.
-
In eukaryotic organisms, the cyclic AMP–protein kinase A (cAMP–PKA) and TOR pathways transmit nutrient-derived signals to regulate a myriad of common targets that control complex translational and transcriptional programmes to coordinate nutrient availability with cell growth and differentiation.
-
Fungi sense gases, such as CO2 and ammonia, to control various cellular responses. Carbonic anhydrase maintains CO2/HCO3− homeostasis and thereby regulates the cAMP–PKA pathway, which in turn controls growth, differentiation and virulence factors of pathogenic fungi. Ammonia gas is an intercolony signalling mediator that has an important role in the growth and survival of multicellular yeast colonies.
-
Opsins, phytochromes and white collar-1 proteins function in light sensing in fungi, with a conserved role for white collar proteins in blue-light sensing in diverse species. The downstream signalling events after light exposure are yet to be fully illuminated.
-
Fungi use evolutionarily conserved signalling pathways, including the p38/Hog1 mitogen-activated protein kinase (MAPK) pathway and the nutrient sensing Tor and cAMP–PKA pathway, to confer cellular responses against various environmental stresses. However, the development of fungal-specific upstream and downstream systems is also evident, as exemplified by the multi-component phosphorelay system.
-
For successful virulence, pathogenic fungi must counteract a plethora of host-specific factors, such as serum and immune cells in animals, and plant hormones, fatty acids and hard mechanical surface in plants. Signalling pathways that are responsible for the fungus–host interaction include the cAMP–PKA pathway and calcineurin pathway, however many aspects of fungal–host interactions remain to be elucidated.
Abstract
All living organisms use numerous signal-transduction systems to sense and respond to their environments and thereby survive and proliferate in a range of biological niches. Molecular dissection of these signalling networks has increased our understanding of these communication processes and provides a platform for therapeutic intervention when these pathways malfunction in disease states, including infection. Owing to the expanding availability of sequenced genomes, a wealth of genetic and molecular tools and the conservation of signalling networks, members of the fungal kingdom serve as excellent model systems for more complex, multicellular organisms. Here, we review recent progress in our understanding of how fungal-signalling circuits operate at the molecular level to sense and respond to a plethora of environmental cues.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bardwell, L. A walk-through of the yeast mating pheromone response pathway. Peptides 25, 1465–1476 (2004).
Han, K. H., Seo, J. A. & Yu, J. H. A putative G protein-coupled receptor negatively controls sexual development in Aspergillus nidulans. Mol. Microbiol. 51, 1333–1345 (2004).
Lemaire, K., Van de Velde, S., Van Dijck, P. & Thevelein, J. M. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol. Cell 16, 293–299 (2004). A comprehensive study that describes how Gpr1 senses different sugars in S. cerevisiae.
Lorenz, M. C. et al. The G protein-coupled receptor Gpr1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics 154, 609–622 (2000).
Maidan, M. M. et al. The G protein-coupled receptor Gpr1 and the Gα protein Gpa2 act through the cAMP–protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16, 1971–1986 (2005). The first report in fungi that a GPCR functions as an extracellular amino-acid sensor and is involved in cAMP–PKA signal activation.
Miwa, T. et al. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3, 919–931 (2004).
Fowler, T. J., DeSimone, S. M., Mitton, M. F., Kurjan, J. & Raper, C. A. Multiple sex pheromones and receptors of a mushroom-producing fungus elicit mating in yeast. Mol. Biol. Cell 10, 2559–2572 (1999).
Versele, M., Lemaire, K. & Thevelein, J. M. Sex and sugar in yeast: two distinct GPCR systems. EMBO Rep. 2, 574–579 (2001).
Tanaka, K., Davey, J., Imai, Y. & Yamamoto, M. Schizosaccharomyces pombe map3+ encodes the putative M-factor receptor. Mol. Cell. Biol. 13, 80–88 (1993).
Seo, J. A., Han, K. H. & Yu, J. H. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53, 1611–1623 (2004).
Kim, H. & Borkovich, K. A. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52, 1781–98 (2004).
Bölker, M., Urban, M. & Kahmann, R. The a mating type locus of U. maydis specifies cell signaling components. Cell 68, 441–450 (1992).
Chung, S. et al. Molecular analysis of CPRα, a MATα-specific pheromone receptor gene of Cryptococcus neoformans. Eukaryot. Cell 1, 432–439 (2002).
Kothe, E., Gola, S. & Wendland, J. Evolution of multispecific mating-type alleles for pheromone perception in the homobasidiomycete fungi. Curr. Genet. 42, 268–275 (2003).
Brown, A. J. & Casselton, L. A. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 17, 393–400 (2001).
Nelson, G. et al. Mammalian sweet taste receptors. Cell 106, 381–390 (2001).
Xue, Y., Batlle, M. & Hirsch, J. P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 17, 1996–2007 (1998).
Welton, R. M. & Hoffman, C. S. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ and the git3 putative glucose receptor. Genetics 156, 513–521 (2000).
Forsberg, H. & Ljungdahl, P. O. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr. Genet. 40, 91–109 (2001).
Brown, V., Sexton, J. A. & Johnston, M. A glucose sensor in Candida albicans. Eukaryot. Cell 5, 1726–1737 (2006).
Madi, L., McBride, S. A., Bailey, L. A. & Ebbole, D. J. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics 146, 499–508 (1997).
Didion, T., Regenberg, B., Jorgensen, M. U., Kielland-Brandt, M. C. & Andersen, H. A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol. 27, 643–650 (1998).
Klasson, H., Fink, G. R. & Ljungdahl, P. O. Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol. 19, 5405–5416 (1999).
Wu, B. et al. Competitive intra- and extracellular nutrient sensing by the transporter homologue Ssy1p. J. Cell Biol. 173, 327–331 (2006).
Andreasson, C., Heessen, S. & Ljungdahl, P. O. Regulation of transcription factor latency by receptor-activated proteolysis. Genes Dev. 20, 1563–1568 (2006).
Martinez, P. & Ljungdahl, P. O. Divergence of Stp1 and Stp2 transcription factors in Candida albicans places virulence factors required for proper nutrient acquisition under amino acid control. Mol. Cell. Biol. 25, 9435–9446 (2005).
Brega, E., Zufferey, R. & Mamoun, C. B. Candida albicans Csy1p is a nutrient sensor important for activation of amino acid uptake and hyphal morphogenesis. Eukaryot. Cell 3, 135–143 (2004).
Thevelein, J. M. et al. Nutrient sensing systems for rapid activation of the protein kinase A pathway in yeast. Biochem. Soc. Trans. 33, 253–256 (2005).
Xue, C., Bahn, Y. S., Cox, G. M. & Heitman, J. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP–PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17, 667–679 (2006).
Lorenz, M. C. & Heitman, J. The Mep2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 17, 1236–1247 (1998).
Smith, D. G., Garcia-Pedrajas, M. D., Gold, S. E. & Perlin, M. H. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol. Microbiol. 50, 259–275 (2003).
Biswas, K. & Morschhäuser, J. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol. Microbiol. 56, 649–669 (2005).
Marini, A. M., Boeckstaens, M., Benjelloun, F., Cherif-Zahar, B. & Andre, B. Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr. Genet. 49, 364–374 (2006).
Van Nuland, A. et al. Ammonium permease-based sensing mechanism for rapid ammonium activation of the protein kinase A pathway in yeast. Mol. Microbiol. 59, 1485–1505 (2006).
Khademi, S. et al. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305, 1587–1594 (2004). The first structure of a member of the Amt/Mep/Rh family of proteins that is consistent with these permeases translocating ammonia gas rather than the ammonium ion.
Lin, Y., Cao, Z. & Mo, Y. Molecular dynamics simulations on the Escherichia coli ammonia channel protein AmtB: mechanism of ammonia/ammonium transport. J. Am. Chem. Soc. 128, 10876–10884 (2006).
Andrade, S. L., Dickmanns, A., Ficner, R. & Einsle, O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl Acad. Sci. USA 102, 14994–14999 (2005).
Marini, A. M., Soussi-Boudekou, S., Vissers, S. & Andre, B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 4282–4293 (1997).
Palková, Z. et al. Ammonia mediates communication between yeast colonies. Nature 390, 532–536 (1997).
Mouillon, J. M. & Persson, B. L. New aspects on phosphate sensing and signalling in Saccharomyces cerevisiae. FEMS Yeast Res. 6, 171–176 (2006).
Heitman, J., Movva, N. R. & Hall, M. N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909 (1991). Discovery of rapamycin-resistant mutants in S. cerevisiae that led to the identification of the Tor proteins.
Loewith, R. et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10, 457–468 (2002). Identification of the TORC1 and TORC2 complexes in S. cerevisiae.
Wedaman, K. P. et al. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 1204–1220 (2003).
Araki, T., Uesono, Y., Oguchi, T. & Toh, E. A. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet. Syst. 80, 325–343 (2005).
Wang, H. & Jiang, Y. The Tap42-protein phosphatase type 2A catalytic subunit complex is required for cell cycle-dependent distribution of actin in yeast. Mol. Cell. Biol. 23, 3116–3125 (2003).
van Slegtenhorst, M., Carr, E., Stoyanova, R., Kruger, W. D. & Henske, E. P. tsc 1+ and tsc2 + regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J. Biol. Chem. 279, 12706–12713 (2004).
Li, Y., Corradetti, M. N., Inoki, K. & Guan, K. L. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29, 32–38 (2004).
Chen, E. J. & Kaiser, C. A. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J. Cell Biol. 161, 333–347 (2003).
Dubouloz, F., Deloche, O., Wanke, V., Cameroni, E. & De Virgilio, C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19, 15–26 (2005). Identification of the EGO complex and demonstration of its involvement in regulating microautophagy together with TORC1. See also reference 50.
Gao, M. & Kaiser, C. A. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nature Cell Biol. 8, 657–667 (2006).
Cardenas, M. E., Cutler, N. S., Lorenz, M. C., Di Como, C. J. & Heitman, J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 13, 3271–3279 (1999). Characterization of the transcriptional response regulated by the TOR pathway in S. cerevisiae . See also references 52–56.
Hardwick, J. S., Kuruvilla, F. G., Tong, J. K., Shamji, A. F. & Schreiber, S. L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl Acad. Sci. USA 96, 14866–14870 (1999).
Powers, T. & Walter, P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10, 987–1000 (1999).
Zaragoza, D., Ghavidel, A., Heitman, J. & Schultz, M. C. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18, 4463–4470 (1998).
Beck, T. & Hall, M. N. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689–692 (1999).
Bertram, P. G. et al. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275, 35727–35733 (2000).
Komeili, A., Wedaman, K. P., O'Shea, E. K. & Powers, T. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151, 863–878 (2000).
Rohde, J. R. & Cardenas, M. E. The Tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23, 629–635 (2003).
Jorgensen, P. et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18, 2491–2505 (2004).
Marion, R. M. et al. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl Acad. Sci. USA 101, 14315–14322 (2004).
Martin, D. E., Soulard, A. & Hall, M. N. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119, 969–979 (2004).
Schawalder, S. B. et al. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432, 1058–1061 (2004).
Rudra, D., Zhao, Y. & Warner, J. R. Central role of Ifh1p–Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 24, 533–542 (2005).
Wade, J. T., Hall, D. B. & Struhl, K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432, 1054–1058 (2004).
Li, H., Tsang, C. K., Watkins, M., Bertram, P. G. & Zheng, X. F. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442, 1058–1061 (2006). Shows that TORC1 is associated to rRNA promoters and thereby regulates Pol I-dependent transcription.
Di Como, C. J. & Arndt, K. T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10, 1904–1916 (1996).
Rohde, J. R. et al. TOR controls transcriptional and translational programs via Sap–Sit4 protein phosphatase signaling effectors. Mol. Cell. Biol. 24, 8332–8341 (2004).
Cherkasova, V. A. & Hinnebusch, A. G. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 17, 859–872 (2003).
Marbach, I., Licht, R., Frohnmeyer, H. & Engelberg, D. Gcn2 mediates Gcn4 activation in response to glucose stimulation or UV radiation not via GCN4 translation. J. Biol. Chem. 276, 16944–16951 (2001).
Gimeno, C. J., Ljungdahl, P. O., Styles, C. A. & Fink, G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68, 1077–1090 (1992).
Pan, X. & Heitman, J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 4874–4887 (1999).
Cutler, N. S., Pan, X., Heitman, J. & Cardenas, M. E. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell 12, 4103–4113 (2001).
Schmidt, A., Bickle, M., Beck, T. & Hall, M. N. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88, 531–542 (1997).
Noda, T. & Ohsumi, Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–2966 (1998).
Budovskaya, Y. V., Stephan, J. S., Reggiori, F., Klionsky, D. J. & Herman, P. K. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279, 20663–20671 (2004).
Schmelzle, T., Beck, T., Martin, D. E. & Hall, M. N. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24, 338–351 (2004).
Zurita-Martinez, S. A. & Cardenas, M. E. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot. Cell 4, 63–71 (2005).
Chen, J. C. & Powers, T. Coordinate regulation of multiple and distinct biosynthetic pathways by TOR and PKA kinases in S. cerevisiae. Curr. Genet. 49, 281–293 (2006).
Granger, D. L., Perfect, J. R. & Durack, D. T. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 76, 508–516 (1985).
Zaragoza, O., Fries, B. C. & Casadevall, A. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2 . Infect. Immun. 71, 6155–6164 (2003).
Mock, R. C., Pollack, J. H. & Hashimoto, T. Carbon dioxide induces endotrophic germ tube formation in Candida albicans. Can. J. Microbiol. 36, 249–253 (1990).
Klengel, T. et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr. Biol. 15, 2021–2026 (2005). Shows that physiological CO 2 concentrations (5%) are sensed by AC to trigger morphological transitions of C. albicans.
Bahn, Y. S., Cox, G. M., Perfect, J. R. & Heitman, J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr. Biol. 15, 2013–2020 (2005). The first study to show the role of fungal CA for growth, virulence and differentiation, using C. neoformans as a model system.
Mogensen, E. G. et al. Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1. Eukaryot. Cell 5, 103–111 (2006).
Bahn, Y. S., Staab, J. & Sundstrom, P. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol. Microbiol. 50, 391–409 (2003).
Bahn, Y. S. & Sundstrom, P. CAP1, an adenylate cyclase-associated protein gene, regulates bud–hypha transitions, filamentous growth and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183, 3211–3223 (2001).
Rocha, C. R. et al. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12, 3631–3643 (2001).
Alspaugh, J. A., Perfect, J. R. & Heitman, J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 11, 3206–3217 (1997).
Chen, Y. et al. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science 289, 625–628 (2000).
Zikánová, B., Kuthan, M., Ricicová, M., Forstová, J. & Palková, Z. Amino acids control ammonia pulses in yeast colonies. Biochem. Biophys. Res. Commun. 294, 962–967 (2002).
Palková, Z. et al. Ammonia pulses and metabolic oscillations guide yeast colony development. Mol. Biol. Cell 13, 3901–3914 (2002).
Váchová, L. et al. Sok2p transcription factor is involved in adaptive program relevant for long term survival of Saccharomyces cerevisiae colonies. J. Biol. Chem. 279, 37973–37981 (2004).
Váchová, L. & Palková, Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 169, 711–717 (2005).
Marsh, P. B., Taylor, E. E. & Bassler, L. M. A guide to the literature on certain effects of light on fungi: reproduction, morphology, pigmentation, and phototropic phenomena. Plant Dis. Rep. Suppl. 261, 251–312 (1959).
Sharma, A. K., Spudich, J. L. & Doolittle, W. F. Microbial rhodopsins: functional versatility and genetic mobility. Trends Microbiol. 14, 463–469 (2006).
Bieszke, J. A. et al. The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc. Natl Acad. Sci. USA 96, 8034–8039 (1999).
Bieszke, J. A., Spudich, E. N., Scott, K. L., Borkovich, K. A. & Spudich, J. L. A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 38, 14138–14145 (1999).
Waschuk, S. A., Bezerra, A. G., Jr, Shi, L. & Brown, L. S. Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote. Proc. Natl Acad. Sci. USA 102, 6879–6883 (2005). Demonstration of the first light-driven proton pump in a eukaryotic organism.
Sumii, M., Furutani, Y., Waschuk, S. A., Brown, L. S. & Kandori, H. Strongly hydrogen-bonded water molecule present near the retinal chromophore of Leptosphaeria rhodopsin, the bacteriorhodopsin-like proton pump from a eukaryote. Biochemistry 44, 15159–15166 (2005).
Saranak, J. & Foster, K. W. Rhodopsin guides fungal phototaxis. Nature 387, 465–466 (1997).
Saranak, J. & Foster, K. W. Photoreceptor for curling behavior in Peranema trichophorum and evolution of eukaryotic rhodopsins. Eukaryot. Cell 4, 1605–1612 (2005).
Blumenstein, A. et al. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15, 1833–1838 (2005). The first report of phytochrome function in the fungal kingdom.
Ballario, P. et al. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15, 1650–1657 (1996).
Liu, Y. & Bell-Pedersen, D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot. Cell 5, 1184–1193 (2006).
Dunlap, J. C. Proteins in the Neurospora circadian clockworks. J. Biol. Chem. 281, 28489–28493 (2006).
Idnurm, A. & Heitman, J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 3, e95 (2005).
Terashima, K., Yuki, K., Muraguchi, H., Akiyama, M. & Kamada, T. The dst1 gene involved in mushroom photomorphogenesis of Coprinus cinereus encodes a putative photoreceptor for blue light. Genetics 171, 101–108 (2005).
Lu, Y. K., Sun, K. H. & Shen, W. C. Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans. Mol. Microbiol. 56, 480–491 (2005).
Idnurm, A. et al. The Phycomyces madA gene encodes a blue-light photoreceptor for phototropism and other light responses. Proc. Natl Acad. Sci. USA 103, 4546–4551 (2006).
Silva, F., Torres-Martínez, S. & Garre, G. Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol. Microbiol. 61, 1023–1037 (2006). Together with Phycomyces , the discoveries in Mucor extend the role of WC-1 proteins in photosensing to include the zygomycetes and show that these fungi contain up to three copies of wc-1 in their genomes derived from recent gene duplications.
Casas-Flores, S., Rios-Momberg, M., Bibbins, M., Ponce-Noyola, P. & Herrera-Estrella, A. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 150, 3561–3569 (2004).
Lee, K. et al. Light regulation of asexual development in the rice blast fungus Magnaporthe grisea. Fungal Genet. Biol. 43, 694–709 (2006).
He, Q. et al. White collar-1, a DNA binding transcription factor and a light sensor. Science 297, 840–843 (2002). Describes the purification of WC-1 from N. crassa and demonstrates that WC-1 contains an associated flavin chromophore.
Schwerdtfeger, C. & Linden, H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22, 4846–4855 (2003).
Heintzen, C., Loros, J. J. & Dunlap, J. C. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104, 453–464 (2001).
Schmoll, M., Franchi, L. & Kubicek, C. P. Envoy, a PAS/LOV domain protein of Hypocrea jecorina (anamorph Trichoderma reesei), modulates cellulase gene transcription in response to light. Eukaryot. Cell 4, 1998–2007 (2005).
Casas-Flores, S. et al. Cross talk between a fungal blue-light perception system and the cyclic AMP signaling pathway. Eukaryot. Cell 5, 499–506 (2006).
Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372 (2002).
Bilsland, E., Molin, C., Swaminathan, S., Ramne, A. & Sunnerhagen, P. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 53, 1743–1756 (2004).
Haghnazari, E. & Heyer, W. D. The Hog1 MAP kinase pathway and the Mec1 DNA damage checkpoint pathway independently control the cellular responses to hydrogen peroxide. DNA Repair (Amst.) 3, 769–776 (2004).
Roman, E., Nombela, C. & Pla, J. The Sho1 adaptor protein links oxidative stress to morphogenesis and cell wall biosynthesis in the fungal pathogen Candida albicans. Mol. Cell. Biol. 25, 10611–10627 (2005). Shows that the C. albicans Sho1 protein has an important role in oxidative stress, cell-wall biogenesis and morphology, which is mainly independent of the MAPK Hog1.
O'Rourke, S. M. & Herskowitz, I. A third osmosensing branch in Saccharomyces cerevisiae requires the Msb2 protein and functions in parallel with the Sho1 branch. Mol. Cell. Biol. 22, 4739–4749 (2002).
Degols, G., Shiozaki, K. & Russell, P. Activation and regulation of the Spc1 stress-activated protein kinase in Schizosaccharomyces pombe. Mol. Cell. Biol. 16, 2870–2877 (1996).
Alonso-Monge, R. et al. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot. Cell 2, 351–361 (2003).
Smith, D. A., Nicholls, S., Morgan, B. A., Brown, A. J. & Quinn, J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol. Biol. Cell 15, 4179–4190 (2004).
Shiozaki, K. & Russell, P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10, 2276–2288 (1996).
Alonso-Monge, R. et al. Role of the mitogen-activated protein kinase Hog1p in morphogenesis and virulence of Candida albicans. J. Bacteriol. 181, 3058–3068 (1999).
Kawasaki, L., Sanchez, O., Shiozaki, K. & Aguirre, J. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 45, 1153–1163 (2002).
Dixon, K. P., Xu, J. R., Smirnoff, N. & Talbot, N. J. Independent signaling pathways regulate cellular turgor during hyperosmotic stress and appressorium-mediated plant infection by Magnaporthe grisea. Plant Cell 11, 2045–2058 (1999).
Bahn, Y. S., Kojima, K., Cox, G. M. & Heitman, J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16, 2285–2300 (2005).
Kojima, K., Bahn, Y. S. & Heitman, J. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology 152, 591–604 (2006).
Kojima, K. et al. Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53, 1785–1796 (2004).
Bahn, Y. S., Kojima, K., Cox, G. M. & Heitman, J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell (2006). A comprehensive study of the two-component systems in a fungus that characterizes six histidine kinases and two response regulators together with the HOG pathway in C. neoformans.
Catlett, N. L., Yoder, O. C. & Turgeon, B. G. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell 2, 1151–1161 (2003).
Kruppa, M. & Calderone, R. Two-component signal transduction in human fungal pathogens. FEMS Yeast Res. 6, 149–159 (2006).
Miller, T. K., Renault, S. & Selitrennikoff, C. P. Molecular dissection of alleles of the osmotic-1 locus of Neurospora crassa. Fungal Genet. Biol. 35, 147–155 (2002).
Buck, V. et al. Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol. Biol. Cell 12, 407–419 (2001).
Yoshimi, A., Kojima, K., Takano, Y. & Tanaka, C. Group III histidine kinase is a positive regulator of Hog1-type mitogen-activated protein kinase in filamentous fungi. Eukaryot. Cell 4, 1820–1828 (2005). Shows that group III histidine kinases are positive regulators of the HOG pathway in C. heterostrophus and N. crassa.
Enjalbert, B., Nantel, A. & Whiteway, M. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol. Biol. Cell 14, 1460–1467 (2003).
Enjalbert, B. et al. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell 17, 1018–1032 (2006). The first global transcript-profiling study to identify downstream targets of Hog1 in pathogenic fungi, using C. albicans as a model system.
Nicholls, S. et al. Msn2- and Msn4-like transcription factors play no obvious roles in the stress responses of the fungal pathogen Candida albicans. Eukaryot. Cell 3, 1111–1123 (2004).
Marchler, G., Schuller, C., Adam, G. & Ruis, H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 12, 1997–2003 (1993).
Smith, A., Ward, M. P. & Garrett, S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17, 3556–3564 (1998).
Gorner, W. et al. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21, 135–144 (2002).
Pascual-Ahuir, A., Posas, F., Serrano, R. & Proft, M. Multiple levels of control regulate the yeast cAMP-response element-binding protein repressor Sko1p in response to stress. J. Biol. Chem. 276, 37373–37378 (2001).
Charizanis, C., Juhnke, H., Krems, B. & Entian, K. D. The oxidative stress response mediated via Pos9/Skn7 is negatively regulated by the Ras/PKA pathway in Saccharomyces cerevisiae. Mol. Gen. Genet. 261, 740–752 (1999).
Blankenship, J. R. & Heitman, J. Calcineurin is required for Candida albicans to survive calcium stress in serum. Infect. Immun. 73, 5767–5774 (2005). Shows that serum sensitivity of C. albicans calcineurin mutants results from endogenous levels of Ca2+ present in serum.
Ferreira, M. E. et al. Functional characterization of the Aspergillus fumigatus calcineurin. Fungal Genet. Biol. 20 Sept 2006 (doi:10.1016/j.fgb.2006.08.004).
Prusty, R., Grisafi, P. & Fink, G. R. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 101, 4153–4157 (2004). The first report that plant hormones have important roles in yeast morphological development.
Klose, J., de Sa, M. M. & Kronstad, J. W. Lipid-induced filamentous growth in Ustilago maydis. Mol. Microbiol. 52, 823–835 (2004).
Kolattukudy, P. E., Rogers, L. M., Li, D., Hwang, C. S. & Flaishman, M. A. Surface signaling in pathogenesis. Proc. Natl Acad. Sci. USA 92, 4080–4087 (1995).
Uchiyama, T. & Okuyama, K. Participation of Oryza sativa leaf wax in appressorium formation by Pyricularia oryzae. Phytochemistry 29, 91–92 (1990).
Champe, S. P. & el-Zayat, A. A. Isolation of a sexual sporulation hormone from Aspergillus nidulans. J. Bacteriol. 171, 3982–3988 (1989).
Calvo, A. M., Gardner, H. W. & Keller, N. P. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 276, 25766–25774 (2001).
Tsitsigiannis, D. I. et al. Aspergillus cyclooxygenase-like enzymes are associated with prostaglandin production and virulence. Infect. Immun. 73, 4548–4559 (2005). This report links fatty-acid metabolism and fungal virulence.
Noverr, M. C. & Huffnagle, G. B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 72, 6206–6210 (2004).
Kung, C. A possible unifying principle for mechanosensation. Nature 436, 647–654 (2005).
Zhou, X. L., Stumpf, M. A., Hoch, H. C. & Kung, C. A mechanosensitive channel in whole cells and in membrane patches of the fungus Uromyces. Science 253, 1415–1417 (1991).
Gustin, M. C., Zhou, X. L., Martinac, B. & Kung, C. A mechanosensitive ion channel in the yeast plasma membrane. Science 242, 762–765 (1988).
Zhou, X. L. et al. The transient receptor potential channel on the yeast vacuole is mechanosensitive. Proc. Natl Acad. Sci. USA 100, 7105–7110 (2003). Shows that there is a mechanosensitive channel in yeast vacuoles.
Zhou, X. L. & Kung, C. A mechanosensitive ion channel in Schizosaccharomyces pombe. EMBO J. 11, 2869–2875 (1992).
Watts, H. J., Véry, A.-A., Perera, T. H., Davies, J. M. & Gow, N. A. Thigmotropism and stretch-activated channels in the pathogenic fungus Candida albicans. Microbiology 144, 689–695 (1998).
Hamer, J. E., Chumley, F. G., Howard, R. J. & Valent, B. A mechanism for surface attachment in spores of a plant pathogenic fungus. Science 239, 288–290 (1988).
Xiao, J. Z., Watanabe, T., Kamakura, T., Ohshimi, A. & Yamaguchi, I. Studies on the cellular differentiation of Magnaporthe grisea. Physicochemical aspects of substratum surfaces in relation to appressorium formation. Physiol. Mol. Plant Pathol. 44, 227–236 (1994).
Aramburu, J., Heitman, J. & Crabtree, G. R. Calcineurin: a central controller of signalling in eukaryotes. EMBO Rep. 5, 343–348 (2004).
Rohde, J., Heitman, J. & Cardenas, M. E. The TOR kinases link nutrient sensing to cell growth. J. Biol. Chem. 276, 9583–9586 (2001).
Schmelzle, T. & Hall, M. N. TOR, a central controller of cell growth. Cell 103, 253–262 (2000).
Acknowledgements
The authors thank F. A. Mühlschlegel for providing pictures of C. albicans filamentation. This work was supported by the Soongsil University Research Fund to Y-S.B. and R01 grants from the NIAID/NIH to J.H. and NCI/NIH to M.E.C.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
Glossary
- Mating type
-
A strain or clone or other isolate made up of organisms (such as certain fungi or protozoans) that are usually incapable of sexual reproduction with one another but capable of such reproduction with members of other strains of the same organism.
- Clamp cell
-
A bridge-like hyphal connection involved in maintaining the dikaryotic state that forms when cells in dikaryotic hyphae divide.
- Protein kinase A
-
(PKA). A secondary messenger-dependent enzyme that has been implicated in a wide range of cellular processes, including transcription, metabolism, cell-cycle progression and apoptosis.
- Rhesus proteins
-
Mammalian homologues of the Amt/Mep family of proteins that are expressed in many tissues and form part of the rhesus (Rh) blood-group complex.
- Autophagy
-
A degradative pathway elicited by nutrient starvation by which indiscriminate portions of the cytoplasm, including organelles, are engulfed into autophagosomal vesicles for fusion with the vacuole and degradation.
- Restenosis
-
The process whereby intimal hyperplasia occurs to re-occlude a coronary artery and limit cardiac blood flow following cardiac stenting. A common complication that is markedly reduced by using stents impregnated with rapamycin.
- EGO complex
-
A vacuolar membrane-associated multiprotein complex that consists of Ego1, Ego3, Gtr1 and Gtr2. Proposed to function in concert with TORC1 to promote microautophagy in response to amino-acid signals.
- Microautophagy
-
The uptake of cytoplasm at the lysosomal or vacuolar surface. It is thought that this process functions to recycle the vacuolar membrane.
- Photosensory protein
-
A protein with absorbance properties that overlap the wavelength spectrum to which the organism responds. Mutation of its encoding gene should disable this sensing ability.
- Chromophore
-
The light-absorbing chemical associated with a photoreceptor protein.
- LOV
-
A domain found in proteins that sense light, oxygen or voltage that physically interacts with a flavin molecule.
- Two-component phosphorelay system
-
First identified in various bacterial systems and subsequently found in lower eukaryotes including fungi. The signalling is achieved by phosphotransfer from a histidine residue in the sensor histidine kinase to an aspartate residue in the response regulator.
- Serum
-
The liquid component of blood that consists of proteins, lipids and many low molecular-weight molecules.
- Appressorium
-
An enlarged fungal filament that is used for penetration through the surface of the host plant.
- Stress regulated element
-
(STRE). A region in the promoter of genes (consensus sequence CCCCT) to which transcription factors bind to mediate stress-induced transcription.
- Calcineurin
-
A serine/threonine-specific protein phosphatase that is activated by calcium–calmodulin.
- Indole-3-acetic acid
-
(IAA). An auxin plant hormone.
- Auxin
-
A class of plant growth substance (often called phytohormones or plant hormones).
Rights and permissions
About this article
Cite this article
Bahn, YS., Xue, C., Idnurm, A. et al. Sensing the environment: lessons from fungi. Nat Rev Microbiol 5, 57–69 (2007). https://doi.org/10.1038/nrmicro1578
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1578
This article is cited by
-
bZIP transcription factors PcYap1 and PcRsmA link oxidative stress response to secondary metabolism and development in Penicillium chrysogenum
Microbial Cell Factories (2022)
-
Oxidative stress response pathways in fungi
Cellular and Molecular Life Sciences (2022)
-
Stress response capacity analysis during aging and possible new insights into aging studies
Current Genetics (2021)