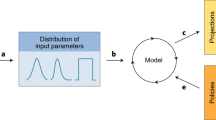
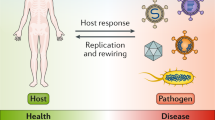
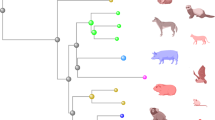
Abstract
Models occupy an essential position in the study of infectious disease as a result of the ethical problems of exposing humans to potentially lethal agents. Deliberately induced infections in well-defined animal models provide much useful information about disease processes in an approximation of their natural context. Despite this, animal models are not the natural disease process, and recent experimental advances show, perhaps not unsurprisingly, that there are large differences between natural infections and animal models. Focusing on mouse models of bacterial pathogens, we discuss some of these discrepancies and suggest ways of improving model systems in the future.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Anon. Scaling up the response to infectious disease [online] <http://www.who.int/infectious-disease-report/2002> (2002).
Anderson, R. M. & May, R. M. Infectious Diseases of Humans: Dynamics and Control (Oxford University Press, Oxford, 1992).
Matthews, L. & Woolhouse, M. New approaches to quantifying the spread of infection. Nature Rev. Microbiol. 3, 529–536 (2005).
Haque, A. et al. Early interactions of Salmonella enterica serovar Typhimurium with human small intestinal epithelial explants. Gut 53, 1424–1430 (2004).
Gravato-Nobre, M. J. & Hodgkin, J. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 7, 741–751 (2005).
Garcia-Lara, J., Needham, A. J. & Foster, S. J. Invertebrates as animal models for Staphylococcus aureus pathogenesis: a window into host–pathogen interaction. FEMS Immunol. Med. Microbiol. 43, 311–323 (2005).
Pradel, E. & Ewbank, J. J. Genetic models in pathogenesis. Annu. Rev. Genet. 38, 347–363 (2004).
Steinert, M., Leippe, M. & Roeder, T. Surrogate hosts: protozoa and invertebrates as models for studying pathogen–host interactions. Int. J. Med. Microbiol. 293, 321–332 (2003).
Sifri, C. D., Begun, J. & Ausubel, F. M. The worm has turned — microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13, 119–127 (2005).
Miller, J. D. & Neely, M. N. Zebrafish as a model host for streptococcal pathogenesis. Acta Trop. 91, 53–68 (2004).
Steinert, M. & Heuner, K. Dictyostelium as host model for pathogenesis. Cell. Microbiol. 7, 307–314 (2005).
Wade, C. M. & Daly, M. J. Genetic variation in laboratory mice. Nature Genet. 37, 1175–1180 (2005).
Committee for laboratory animal care and use. The numbers of live animals used in experiments in 2001— results of a survey. Exp. Anim. 52, 143 (2003).
The Commission to the Council and the European Parliament. Fourth report on the statistics on the number of animals used for experimental and other scientific purposes in the member states of the European Union [online] <http://europa.eu.int/comm/environment/chemicals/lab_animals/pdf/ com_2005_7_en.pdf> (2005).
Matsuda, Y. Recent trends in the number of laboratory animals used in Japan ATLA 32 (suppl. 1), 299–301 (2004)
Home Office. Statistics of Scientific Procedures on Living Animals Great Britain 2003 (HMSO, London, 2004).
Vazquez-Boland, J. A. et al. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640 (2001).
Okamoto, M., Nakane, A. & Minagawa, T. Host resistance to an intragastric infection with Listeria monocytogenes in mice depends on cellular immunity and intestinal bacterial flora. Infect. Immun. 62, 3080–3085 (1994).
Marco, A. J. et al. A microbiological, histopathological and immunohistological study of the intragastric inoculation of Listeria monocytogenes in mice. J. Comp. Pathol. 107, 1–9 (1992).
Conlan, J. W. & North, R. J. Neutrophil-mediated dissolution of infected host cells as a defense strategy against a facultative intracellular bacterium. J. Exp. Med. 174, 741–744 (1991).
Mengaud, J., Ohayon, H., Gounon, P., Mege, R. M. & Cossart, P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84, 923–932 (1996).
Lecuit, M. et al. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18, 3956–3963 (1999).
Lecuit, M. et al. A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292, 1722–1725 (2001).
Schlievert, P. M., Assimacopoulos, A. P. & Cleary, P. P. Severe invasive group A streptococcal disease: clinical description and mechanisms of pathogenesis. J. Lab. Clin. Med. 127, 13–22 (1996).
Sriskandan, S., Unnikrishnan, M., Krausz, T. & Cohen, J. Molecular analysis of the role of streptococcal pyrogenic exotoxin A (SPEA) in invasive soft-tissue infection resulting from Streptococcus pyogenes. Mol. Microbiol. 33, 778–790 (1999).
Sriskandan, S. et al. Enhanced susceptibility to superantigen-associated streptococcal sepsis in human leukocyte antigen-DQ transgenic mice. J. Infect. Dis. 184, 166–173 (2001).
Scaramuzzino, D. A., McNiff, J. M. & Bessen, D. E. Humanized in vivo model for streptococcal impetigo. Infect. Immun. 68, 2880–2887 (2000).
Sun, H. et al. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305, 1283–1286 (2004).
Garmendia, J., Frankel, G. & Crepin, V. F. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73, 2573–2585 (2005).
Mundy, R., Macdonald, T. T., Dougan, G., Frankel, G. & Wiles, S. Citrobacter rodentium of mice and man. Cell. Microbiol. 7, 1697–1706 (2005).
Chen, H. D. & Frankel, G. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 29, 83–98 (2005).
Thorpe, C. M. Shiga toxin-producing Escherichia coli infection. Clin. Infect. Dis. 38, 1298–1303 (2004).
Truman, R. W. & Krahenbuhl, J. L. Viable M. leprae as a research reagent. Int. J. Lepr. Other Mycobact. Dis. 69, 1–12 (2001).
Truman, R. Leprosy in wild armadillos. Lepr. Rev. 76, 198–208 (2005).
Dean, G. S. et al. Minimum infective dose of Mycobacterium bovis in cattle. Infect. Immun. 73, 6467–6471 (2005).
Buddle, B. M., Wedlock, D. N., Denis, M. & Skinner, M. A. Identification of immune response correlates for protection against bovine tuberculosis. Vet. Immunol. Immunopathol. 108, 45–51 (2005).
Head, N. E. & Yu, H. Cross-sectional analysis of clinical and environmental isolates of Pseudomonas aeruginosa: biofilm formation, virulence, and genome diversity. Infect. Immun. 72, 133–144 (2004).
Cooper, T. F., Rozen, D. E. & Lenski, R. E. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl Acad. Sci. USA 100, 1072–1077 (2003).
Somerville, G. A. et al. Synthesis and deformylation of Staphylococcus aureus δ-toxin are linked to tricarboxylic acid cycle activity. J. Bacteriol. 185, 6686–6694 (2003).
Welch, R. A. et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl Acad. Sci. USA 99, 17020–17024 (2002).
Andersen, P. & Doherty, T. M. The success and failure of BCG — implications for a novel tuberculosis vaccine. Nature Rev. Microbiol. 3, 656–662 (2005).
Merrell, D. S. et al. Host-induced epidemic spread of the cholera bacterium. Nature 417, 642–645 (2002).
Wiles, S., Dougan, G. & Frankel, G. Emergence of a 'hyperinfectious' bacterial state after passage of Citrobacter rodentium through the host gastrointestinal tract. Cell. Microbiol. 7, 1163–1172 (2005).
Alam, A. et al. Hyperinfectivity of human-passaged Vibrio cholerae can be modeled by growth in the infant mouse. Infect. Immun. 73, 6674–6679 (2005).
Rasmussen, M. A. et al. Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect. Immun. 73, 4668–4675 (2005).
Cirillo, J. D., Falkow, S. & Tompkins, L. S. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62, 3254–3261 (1994).
Cirillo, J. D., Falkow, S., Tompkins, L. S. & Bermudez, L. E. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect. Immun. 65, 3759–3767 (1997).
Perry, R. D. & Fetherston, J. D. Yersinia pestis — etiologic agent of plague. Clin. Microbiol. Rev. 10, 35–66 (1997).
Jarrett, C. O. et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190, 783–792 (2004).
Jarrett, C. O., Sebbane, F., Adamovicz, J. J., Andrews, G. P. & Hinnebusch, B. J. Flea-borne transmission model to evaluate vaccine efficacy against naturally acquired bubonic plague. Infect. Immun. 72, 2052–2056 (2004).
Yoshimatsu, T., Shirai, M., Nagata, K., Okita, K. & Nakazawa, T. Transmission of Helicobacter pylori from challenged to nonchallenged nude mice kept in a single cage. Dig. Dis. Sci. 45, 1747–1753 (2000).
Wijburg, O. L. et al. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J. Exp. Med. 203, 21–26 (2006).
Lipsitch, M. & Moxon, E. R. Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol. 5, 31–37 (1997).
Ebert, D. in Evolution in Health and Disease (ed. Stearns, S. C.) 161–172 (Oxford University Press, Oxford, 1999).
Watson, D. A. & Musher, D. M. A brief history of the pneumococcus in biomedical research. Semin. Respir. Infect. 14, 198–208 (1999).
Hanage, W. P. et al. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect. Immun. 73, 431–435 (2005).
Brueggemann, A. B. et al. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187, 1424–1432 (2003).
Hanage, W. P. et al. Using multilocus sequence data to define the pneumococcus. J. Bacteriol. 187, 6223–6230 (2005).
Yuk, M. H., Harvill, E. T., Cotter, P. A. & Miller, J. F. Modulation of host immune responses, induction of apoptosis and inhibition of NF-κB activation by the Bordetella type III secretion system. Mol. Microbiol. 35, 991–1004 (2000).
Mouslim, C., Hilbert, F., Huang, H. & Groisman, E. A. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 45, 1019–1027 (2002).
Acknowledgements
The authors gratefully acknowledge the generous support of the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Related links
DATABASES
Entrez Genome Project
Salmonella enterica serovar Typhimurium
FURTHER INFORMATION
Rights and permissions
About this article
Cite this article
Wiles, S., Hanage, W., Frankel, G. et al. Modelling infectious disease — time to think outside the box?. Nat Rev Microbiol 4, 307–312 (2006). https://doi.org/10.1038/nrmicro1386
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1386
This article is cited by
-
The contribution of bovines to human health against viral infections
Environmental Science and Pollution Research (2021)
-
Model or meal? Farm animal populations as models for infectious diseases of humans
Nature Reviews Microbiology (2010)
-
Cholera transmission: the host, pathogen and bacteriophage dynamic
Nature Reviews Microbiology (2009)
-
Identifying and hurdling obstacles to translational research
Nature Reviews Immunology (2007)