Key Points
-
This review provides an overview of the mycoviruses that are associated with the attenuation of fungal pathogenesis that is known as hypovirulence.
-
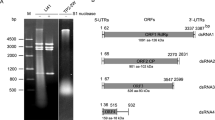
The taxonomy and genome organization of hypoviruses are outlined. These viruses are in the taxonomic family Hypoviridae, which cause hypovirulence of the chestnut blight fungus Cryphonectria parasitica.
-
The development of hypovirus reverse genetics systems is described.
-
Hypovirus diversity has been exploited to identify symptom determinants and modulate fungal–plant pathogenic interactions.
-
Potentially, mycoviruses could be used to explore the origins, evolution and functions of RNA silencing in fungi. These viruses could also be used to probe the signal transduction pathways that underpin fungal pathogenesis.
-
Hypovirus-mediated modulation of the host transcriptome has been analysed using microarrays, which has confirmed hypovirus-mediated alterations of host functions including G-protein signalling, and has revealed a link between mitochondrial and viral hypovirulence.
-
Finally, the prospects for engineering hypoviruses to improve their suitability for biological control of fungal diseases of plants are discussed.
Abstract
Whereas most mycoviruses lead 'secret lives', some reduce the ability of their fungal hosts to cause disease in plants. This property, known as hypovirulence, has attracted attention owing to the importance of fungal diseases in agriculture and the limited strategies that are available for the control of these diseases. Using one pathogen to control another is appealing, both intellectually and ecologically. The recent development of an infectious cDNA-based reverse genetics system for members of the Hypoviridae mycovirus family has enabled the analysis of basic aspects of this fascinating virus–fungus–plant interaction, including virus–host interactions, the mechanisms underlying fungal pathogenesis, fungal signalling pathways and the evolution of RNA silencing. Such systems also provide a means for engineering mycoviruses for enhanced biocontrol potential.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Buck, K. W. in Fungal Virology (ed. Buck, K. W.) 2–84 (CRC, Boca Raton, 1986).
Anagnostakis, S. L. Biological control of chestnut blight. Science 215, 466–471 (1982). One of the first reviews of the use of hypovirulence as a biological control strategy and one of the best descriptions of the North American chestnut blight epidemic.
Brasier, C. M., in The Elms: Breeding, Conservation and Disease Management (ed. Dunn, C. P.) 61–72 (Kluwer, Boston, 2000). Excellent source for information on Dutch elm disease and the hypovirulence-associated viruses of O. ulmi and O. novo-ulmi.
Ghabrial, S. A., Soldevila, A. I. & Havens, W. M. in Molecular Biology of double-stranded RNA: Concepts and Applications in Agriculture, Forestry and Medicine (ed. Tavantzis, S.) 213–236 (CRC, Boca Raton, 2002).
Wei, C. Z., Osaki, H., Iwanami, T., Matsumoto, N. & Ohtsu, Y. Molecular characterization of dsRNA segments 2 and 5 and electron microscopy of a novel reovirus from a hypovirulent isolate, W370, of the plant pathogen Rosellina necatrix. J. Gen. Virol. 84, 2431–2437 (2003).
Suzuki, N., Supyani, S., Maruyama, K. & Hillman, B. I. Complete genome sequence of Mycoreovirus-1/Cp9B21, a member of a novel genus within the family Reoviridae, isolated from the chestnut blight fungus Cryphonectria parasitica. J. Gen. Virol. 85, 3437–3448 (2004).
Wickner, R. B., Wang, C. C. & Patterson, J. L. in Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses (eds Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U. & Ball, L. A.) 571–580 (Elsevier, London, 2005).
Ghabrial, S. A., Jiang, D. & Caston, J. R. in Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses (eds Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U. & Ball, L. A.) 591–595 (Elsevier, London, 2005).
Mertens, P. P. C., Hillman, B. I., & Suzuki, N. in Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses (eds Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U. & Ball, L. A.) (in the press) (Elsevier, London, 2005).
Wickner, R. B., Esteban, R. & Hillman, B. I. in Virus Taxonomy: VIIth report of the International Committee on Taxonomy of Viruses. (eds Van Regenmortel, M. H. et al.) 651–656 (Academic, San Diego, 2000).
Polashock, J. J. & Hillman, B. I. A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc. Natl Acad. Sci. USA 91, 566–571 (1994).
Hong, Y., Dover, S. L., Cole, T. E., Brasier, C. M. & Buck, K. W. Multiple mitochondrial viruses in an isolate of the Dutch elm disease fungus Ophiostoma novo-ulmi. Virology 246, 158–169 (1999).
Deng, F., Xu, R. & Boland, G. J. Hypovirulence-associated double-stranded RNA from Sclerotinia homoeocarpa is conspecific with Ophiostoma novo-ulmi mitovirus 3a-Ld. Phytopathology 93, 1407–1414 (2003).
Cole, T. E., Hong, Y., Brasier, C. M. & Buck, K. W. Detection of an RNA-dependent RNA polymerase in mitochondria from a mitovirus-infected isolate of the Dutch Elm Disease fungus Ophiostoma novo-ulmi. Virology 268, 239–243 (2000).
Shapira, R., Choi, G. H. & Nuss, D. L. Virus-like genetic organization and expression strategy for a double-stranded RNA genetic element associated with biological control of chestnut blight. EMBO J. 10, 731–739 (1991).
Hillman, B. I., Tian, Y., Bedker, P. J. & Brown, M. P. A North American hypovirulent isolate of the chestnut blight fungus with European isolate-related dsRNA. J. Gen. Virol. 73, 681–686 (1992).
Chen, B. & Nuss, D. L. Infectious cDNA clone of hypovirus CHV1/Euro7: a comparative virology approach to investigate virus-mediated hypovirulence of the chestnut blight fungus Cryphonectria parasitica. J. Virol. 73, 985–992 (1999).
Smart, C. D. et al. Cryphonectria hypovirus 3, a virus in the family Hypoviridae with a single open reading frame. Virology 265, 66–73 (1999).
Linder-Basso, D., Dynek, J. & Hillman, B. I. Genome analysis of Cryphonectria hypovirus 4, the most common hypovirus species in North America. Virology (in the press).
Preisig, O., Moleleki, N., Smit, W. A., Wingfield, B. D. & Wingfield, M. J. A novel RNA mycovirus in a hypovirulent isolate of the plant pathogen Diaporthe ambigua. J. Gen. Virol. 81, 3107–3114 (2000).
Lakshman, D. K., Jian, J. & Tavantzis, S. M. A double-stranded RNA element from a hypovirulent strain of Rhizoctonia solani occurs in DNA form and is genetically related to the pentafunctional AROM protein of the shikimate pathway. Proc. Natl Acad. Sci. USA 95, 6425–6429 (1998).
Chu, Y. -M. et al. Double-stranded RNA mycovirus from Fusarium graminearum. Appl. Environ. Microbiol. 68, 2529–2534 (2002).
Buck, K. W. in Molecular Variability of Fungal Pathogens (eds Bridge, P. D., Couteaudier, Y. & Clarkson, J. M.) 53–72 (CAB International, Wallingford, 1998). This review provides an excellent overview of the genome organization and expression strategies of fungal viruses.
Ghabrial, S. A. Origin, adaptation and evolutionary pathways of fungal viruses. Virus Genes 16, 119–131 (1998). Provides perspectives on the origin and evolution of fungal viruses.
Castro, M., Kramer, K., Valdivia, L., Ortiz, S. & Castillo, A. A double-stranded RNA mycovirus confers hypovirulence-associated traits to Botrytis cinerea. FEMS Microbiol. Lett. 228, 87–91 (2003).
Hillman, B. I. & Suzuki, N. Viruses of the chestnut blight fungus. Adv. Virus Res. 63, 423–472 (2004). A recent and comprehensive review of viruses associated with the chestnut blight fungus C. parasitica.
Koonin, E. V., Choi, G. H., Nuss, D. L., Shapira, R. & Carrington, J. C. Evidence for common ancestry of a chestnut blight hypovirulence-associated double-stranded RNA and a group of positive-strand RNA plant viruses. Proc. Natl Acad. Sci. USA 88, 10647–10651 (1991).
Chen, B., Choi, G. H. & Nuss, D. L. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264, 1762–1764 (1994).
Racaniello, V. R. & Baltimore, D. Cloned poliovirus cDNA is infectious in mammalian cells. Science 214, 916–919 (1981).
Ahlquist, P., French, R., Janda, M. & Loesch-Fries, L. S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl Acad. Sci. USA 81, 7066–7070 (1984).
Choi, G. H. & Nuss, D. L. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 257, 800–803 (1992). Describes the construction of an infectious hypovirus cDNA, which established the first mycovirus reverse genetics system.
Moleleki, N., van Heerden, S. W., Wingfield, M. J., Wingfield, B. D. & Preisig, O. Transfection of Diaporthe perjuncta with Diaporthe RNA virus. Appl. Environ. Microbiol. 69, 3952–3956 (2003).
Churchill, A. C. L., Ciufetti, L. M., Hansen, D. R., Van Etten, H. D. & Van Alfen, N. K. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17, 25–31 (1990).
Sasaki, A. et al. Extending chestnut blight hypovirus host range within diaporthales by biolistic delivery of viral cDNA. Mol. Plant. Microb. Interact. 15, 780–789 (2002).
Chen, B., Craven, M. G., Choi, G. H. & Nuss, D. L. cDNA-derived hypovirus RNA in transformed chestnut blight fungus is spliced and trimmed of vector nucleotides. Virology 202, 441–448 (1994).
van Heerden, S. W. et al. Characterization of South African Cryphonectria cubensis isolates infected with a C. parasitica hypovirus. Phytopathology 91, 628–632 (2001).
Elliston, J. E. Characterization of dsRNA-free and dsRNA-containing strains of Endothia parasitica in relation to hypovirulence. Phytopathology 75, 151–158 (1985).
Anagnostakis, S. L. & Day, P. R. Hypovirulence conversion in Endothia parasitica. Phytopathology 69, 1226–1229 (1979).
Anagnostakis, S. L. in The Ecology and Physiology of the Fungal mycelium (eds Jennings, D. H. & Rayner, A. D. M.) 353–366 (Cambridge University Press, Cambridge, 1984).
Peever, T. L., Liu, Y. -C. & Milgroom, M. G. Incidence and diversity of hypoviruses and other double-stranded RNAs occurring in the chestnut blight fungus Cryphonectria parasitica, in China and Japan. Phytopathology 88, 811–17 (1998).
Allemann, C., Hoegger, P., Heiniger, U. & Rigling, D. Genetic variation of Cryphonectria hypoviruses (CHV1) in Europe, assessed using restriction fragment length polymorphism (RFLP) markers. Mol. Ecol. 8, 843–854 (1999).
Chung, P., Bedker, P. J. & Hillman, B. I. Diversity of Cryphonectria parasitica hypovirulence-associated double-stranded RNAs within a chestnut population in New Jersey. Phytopathology 84, 984–990 (1994).
Enebak, S. A., MacDonald, W. L. & Hillman, B. I. Effect of dsRNA associated with isolates of Cryphonectria parasitica from the central Appalachians and their relatedness to other dsRNAs from North America and Europe. Phytopathology 84, 528–534 (1994).
Peever, T. L., Liu, Y. -C. & Milgroom, M. G. Diversity of hypoviruses and other double-stranded RNAs in Cryphonectria parasitica in North America. Phytopathology 87, 1026–1033 (1997).
Chen, B., Geletka, L. M. & Nuss, D. L. Using chimeric hypoviruses to fine-tune the interaction between a pathogenic fungus and its plant host. J. Virol. 74, 7562–7567 (2000).
Suzuki, N., Geletka, L. M. & Nuss, D. L. Essential and dispensible virus-encoded replication elements revealed by efforts to develop hypoviruses as gene expression vectors. J. Virol. 74, 7568–7577 (2000).
Craven, M. G., Pawlyk, D. M., Choi, G. H. & Nuss, D. L. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 67, 6513–6521 (1993).
Suzuki, N., Chen, B. & Nuss, D. L. Mapping of a hypovirus p29 protease symptom determinant domain with sequence similarity to potyvirus HC-Pro protease. J. Virol. 73, 9478–9484 (1999).
Suzuki, N. & Nuss, D. L. The contribution of protein p40 to hypovirus-mediated modulation of fungal host phenotype and viral RNA accumulation. J. Virol. 76, 7747–7759 (2002).
Suzuki, N., Maruyama, K., Moriyama, M. & Nuss, D. L. Hypovirus papain-like protease p29 functions in trans to enhance viral double-stranded RNA accumulation and vertical transmission. J. Virol. 77, 11697–11707 (2003).
Napoli, C., Lemieux, C. & Jorgensen, R. Introduction of a chalocone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289 (1990).
van der Krol, A. R., Mur, L. A., Beld, M., Mol, J. N. & Stuitje, A. R. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to suppression of gene expression. Plant Cell 2, 291–299 (1990).
Romano, N. & Macino, G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 6, 3343–3345 (1992).
Fire, A. RNA-triggered gene silencing. Trends Genet. 15, 358–363 (1999).
Voinnet, O. RNA silencing as a plant immune system against viruses. Trends Genet. 17, 449–459 (2001).
Li, H. W., Li, W. X. & Ding, S. W. Induction and suppression of RNA silencing by an animal virus. Science 296, 1319–1321 (2002).
Silhavy, D. & Burgyan, J. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9, 76–83 (2004).
Choi, G. H., Pawlyk, D. M., & Nuss, D. L. The autocatalytic protease p29 encoded by a hypovirulence-associated virus of the chestnut blight fungus resembles the potyvirus-encoded protease HC-Pro. Virology 183, 747–752 (1991).
Choi, G. H., Shapira, R. & Nuss, D. L. Co-translational autoproteolysis involved in gene expression from a double-stranded RNA genetic element associated with hypovirulence of the chestnut blight fungus. Proc. Natl Acad. Sci. USA 88, 1167–1171 (1991).
Brigneti, G. O. et al. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17, 6739–6746 (1998).
Hannon, G. J. RNA interference. Nature 418, 244–251 (2002).
Kasschau, K. D. et al. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and mRNA function. Dev. Cell. 4, 205–217 (2003).
Catalanotto, C. et al. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24, 2536–2545 (2004).
Chicas, A. & Macino, G. Characteristics of post-transcriptional gene silencing. EMBO Rep. 2, 992–996 (2001).
Cogoni, C. Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55, 381–406 (2001). Includes description of the elements of RNA silencing in fungi.
Dawe, A. L. et al. An ordered collection of expressed sequences from Cryphonectria parasitica and evidence of genomic microsynteny with Neurospora crassa and Magnaporthe grisea. Microbiology 149, 2373–2384 (2003). Emphasizes the concept of using mycoviruses to understand and manage fungal virulence.
Larson, T. G., Choi, G. H. & Nuss, D. L. Regulatory pathways governing modulation of fungal gene expression by a virulence-attenuating mycovirus. EMBO J. 11, 4539–4548 (1992).
Larson, T. G. & Nuss, D. L. Altered transcriptional response to nutrient availability in hypovirus-infected chestnut blight fungus. EMBO J. 13, 5616–5623 (1994).
Park, S. M. et al. Characterization of HOG1 homologue, CpMK1, from Cryphonectria parasitica and evidence for hypovirus-mediated perturbation of its phosphorylation in response to hypertonic stress. Mol. Microbiol. 51, 1267–1277 (2004).
Kim, M. J. et al. Characterization of a fungal protein kinase from Cryphonectria parasitica and its transcriptional upregulation by hypovirus. Mol. Microbiol. 45, 933–941 (2002).
Choi, G. H., Chen, B. & Nuss, D. L. Virus-mediated or transgenic suppression of a G-protein α subunit and attenuation of fungal virulence. Proc. Natl Acad. Sci. USA 92, 305–309 (1995).
Dawe, A. L., Segers, G. C., Allen, T. D., McMains, V. C. & Nuss, D. L. Microarray analysis of Cryphonectria parasitica Gα and Gβγ-signalling pathways reveals extensive modulation by hypovirus infection. Microbiology 150, 4033–4043 (2004).
Gao, S. & Nuss, D. L. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc. Natl Acad. Sci. USA 93, 14122–14127 (1996).
Nuss, D. L. Using hypoviruses to probe and perturb signal transduction processing underlying fungal pathogenesis. Plant Cell 8, 1845–1853 (1996).
Dawe, A. L. & Nuss, D. L. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annu. Rev. Genet. 35, 1–29 (2001).
Allen, T. D., Dawe, A. L. & Nuss, D. L. Use of cDNA microarrays to monitor transcriptional responses of the chestnut blight fungus Cryphonectria parasitica to infection by virulence-attenuating hypoviruses. Eukaryotic Cell 2, 1253–1265 (2003).
Soanes, D. M., Skinner, W., Keon, J., Hargraves, J. & Talbot, N. J. Genomics of phytopathogenic fungi and the development of bioinformatics resources. Mol. Plant Microbe Interact. 15, 421–427 (2002).
Kang, H. -S. et al. Ordered differential display from Cryphonectria parasitica. Plant Pathol. J. 16, 142–146 (2000).
Kasahara, S., Wang, P. & Nuss, D. L. Identification of bdm-1, a gene involved in G-protein β-subunit function and α-subunit accumulation. Proc. Natl Acad. Sci. USA 97, 412–17 (2000).
Yang, Q., Poole, S. I. & Borkovich, K. A. A G-protein β subunit required for sexual and vegetative development and maintenance of normal Gα protein levels in Neurospora crassa. Eukaryotic Cell 1, 378–390 (2002).
Parsley, T. B., Segers, G. C., Nuss, D. L. & Dawe, A. L. Analysis of altered G-protein subunit accumulation in Cryphonectria parasitica reveals a third G α homologue. Curr. Genet. 43, 24–33 (2003).
Monteiro-Vitorello, C. B., Bell, J. A., Fullbright, D. W. & Bertrand, H. A cytoplasmically transmissible hypovirulence phenotype associated with mitochondrial DNA mutations in the chestnut blight fungus Cryphonectria parasitica. Proc. Natl Acad. Sci. USA 92, 5935–5939 (1995).
Fulbright, D. W. A cytoplasmic hypovirulent strain of Endothia parasitica without double-stranded RNA. Phytopathology 75, 1328 (1985).
Mahanti, N., Bertrand, H., Monteiro-Vitorello, C. & Fulbright, D. W. Elevated mitochondrial alternative oxidase activity in dsRNA-free, hypovirulent isolates of Cryphonectria parasitica. Physiol. Mol. Plant Pathol. 42, 455–463 (1993).
Griffiths, A. J. F. Mitochondrial inheritance in filamentous fungi. J. Genet. 75, 403–414 (1996).
Allen, T. D. & Nuss, D. L. Linkage between mitochondrial hypovirulence and viral hypovirulence in the chestnut blight fungus revealed by cDNA microarray analysis. Eukaryotic Cell 3, 1227–1232 (2004).
Burtrand, H. Role of mitochondrial DNA in the senescence and hypovirulence of fungi and potential for plant disease control. Annu. Rev. Phytopathol. 38, 397–422 (2000). Excellent review of the relationship between mitochondrial dysfunction and the attenuation of fungal virulence known as mitochondrial hypovirulence.
Dementhon, K. et al. Rapamycin mimics the incompatibility reaction in the fungus Podospora anserina. Eukaryotic Cell 2, 238–246 (2003).
Dennis, P. et al. Mammalian TOR: a homeostatic ATP sensor. Science 294, 1102–1105 (2001).
Desai, B., Myers, B. & Schreiber, S. FKBP12-rapamycin-associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc. Natl Acad. Sci. USA 99, 4319–4324 (2002).
Biraghi, A. Possible active resistance to Endothia parasitica in Castanea sativa in Rep. Congr. Int. Union For. Res. Org. 11th. 149–157 (International Union of Forest Research Organization, Rome, 1953).
Grente, J. & Berthelay-Sauret, S. in Proceedings of the American Chestnut Symposium, (eds MacDonald, W. L., Cech, F. C., Luchok, J. & Smith, C.) 30–34 (West Virginia University, Morgantown, 1978).
Day, P. R., Dodds, J. A., Elliston, J. E., Jaynes, R. A., & Anagnostakis, S. L. Double-stranded RNA in Endothia parasitica. Phytopathology 68, 1391–1396 (1977).
Heiniger, U., & Rigling, D. Biological control of chestnut blight in Europe. Annu. Rev. Phytopathol. 32, 581–599 (1994). Excellent review of the history and current picture of chestnut blight and hypovirulence in Europe.
Griffin, G. J. Chestnut blight and its control. Hortic. Rev. 8, 291–336 (1986).
MacDonald, W. L. & Fulbright, D. W. Biological control of chestnut blight: use and limitation of transmissible hypovirulence. Plant Dis. 75, 656–661 (1991). This review provides an excellent perspective on the importance of balancing the effects of hypovirus infection on ecological fitness and virulence attenuation for successful hypovirulence-mediated biological control.
Fulbright, D. W., Weidlich, W. H., Haufler, Z., Thomas, C. S. & Paul, C. P. Chestnut blight and recovering American chestnut trees in Michigan. Can. J. Bot. 61, 3144–3171 (1983).
Milgroom, M. G. & Cortesi, P. Biological control of chestnut blight with hypovirulence: a critical analysis. Annu. Rev. Phytopathol. 42, 311–338 (2004). Recent critical assessment of the effectiveness of hypovirulence in the management of chestnut blight.
Cortesi, P. & Milgroom, M. G. Genetics of vegetative incompatibility in Cryphonectria parasitica. Appl. Environ. Microbiol. 64, 2988–2994 (1998).
Liu, Y. -C. & Milgroom, M. G. Correlation between hypovirus transmission and the number of vegetative incompatibility (vic) genes different among isolates from natural populations of Cryphonectria parasitica. Phytopathology 86, 79–86 (1996).
Cortesi, P., McCulloch, C. E., Song, H., Lin, H. & Milgroom, M. G. Genetic control of horizontal virus transmission in the chestnut blight fungus, Cryphonectria parasitica. Genetics 159, 107–118 (2001).
Anagnostakis, S. L. & Kranz, J. Population dynamics of Cryphonectria parasitica in a mixed-hardwood forest in Connecticut. Phytopathology 77, 751–754 (1987).
Bissegger, M., Rigling, D. & Heiniger, U. Population structure and disease development of Cryphonectria parasitica in European chestnut forests in the presence of natural hypovirulence. Phytopathology 87, 51–59 (1997).
Chen, B., Chen, C. -H., Bowman, B. H. & Nuss, D. L. Phenotypic changes associated with wild-type and mutant hypovirus RNA transfection of plant pathogenic fungi phylogenetically related to Cryphonectria parasitica. Phytopathology 86, 301–310 (1996).
Shain, L. & Miller, J. B. Movement of cytoplasmic hypovirulence agents in chestnut blight cankers. Can. J. Bot. 70, 557–561 (1992).
Chen, B., Choi, G. H. & Nuss, D. L. Mitotic stability and nuclear inheritance of integrated viral cDNA in engineered hypovirulent strains of the chestnut blight fungus. EMBO J. 12, 2991–2998 (1993).
Liu, Y. -C., Durrett, R. & Milgroom, M. G. A spatially-structured stochastic model to simulate heterogeneous transmission of viruses in fungal populations. Ecol. Modell. 127, 291–308 (2000).
Anagnostakis, S. L., Chen, B., Geletka, L. M. & Nuss, D. L. Hypovirus transmission to ascospore progeny by field-released transgenic hypovirulent strains of Cryphonectria parasitica. Phytopathology 88, 598–604 (1998).
Root, C., et al. Multiseasonal field release and spermatization of transgenic hypovirulent strains of Cryphonectria parasitica containing cDNA copies of the highly debilitating hypovirus CHV1-EP713. For. Pathol. (in the press).
Gobbin, D., Hoegger, P. J., Heiniger, U. & Rigling, D. Sequence variation and evolution of Cryphonectria hypovirus 1 (CHV1) in Europe. Virus Res. 97, 39–46 (2003).
Graves, A. H. Relative blight resistance in species and hybrids of Castanea. Phytopathology 40, 1125–1131 (1950).
Hebard, F. V. Inheritance of juvenile leaf and stem morphological traits in crosses of Chinese and American chestnut. J. Hered. 85, 440–446 (1994).
Connors, B. J., Laun, N. P., Maynard, C. A. & Powell, W. A. Molecular characterization of a gene encoding a cystatin expressed in the stems of American chestnut (Castanea dentate). Planta 3, 510–514 (2002).
Jian, J. H., Lakshman, D. K. & Tavantzis, S. M. Association of distinct double-stranded RNAs with enhanced or diminished virulence in Rhizoctonia solani infecting potato. Mol. Plant Microbe Interact. 10, 1002–1009 (1997).
Jian, J. H., Lakshman, D. K. & Tavantzis, S. M. A virulence-associated, 6.4-kb, double-stranded RNA from Rhizoctonia solani is phylogenetically related to plant bromoviruses and electron transport enzymes. Mol. Plant Microbe Interact. 11, 601–609 (1998).
Liu, C., Lakshman, D. K. & Tavantzis, S. M. Quinic acid induces hypovirulence and expression of a hypovirulence-associated double-stranded RNA in Rhizoctonia solani. Curr. Genet. 43, 103–111 (2003).
Polashock, J. J., Bedker, P. J. & Hillman, B. I. Movement of a small mitochondrial double-stranded RNA element of Cryphonectria parasitica: ascospore inheritance and implications for mitochondrial recombination. Mol. Gen. Genet. 256, 566–571 (1997).
Shapira, R. & Nuss, D. L. Gene expression by a hypovirulence-associated virus of the chestnut blight fungus involves two papain-like protease activities. J. Biol. Chem. 266, 19419–19425 (1991).
Nuss, D. L., Chen, B., Geletka, L. M., Parsley, T. B. & Suzuki, N. in Molecular Biology of double-stranded RNA: Concepts and Applications in Agriculture, Forestry and Medicine (ed. Tavantzis, S.) 145–163 (CRC, Boca Raton, 2002).
Buck, K. W. & Brasir, C. M. in Molecular Biology of double-stranded RNA: Concepts and Applications in Agriculture, Forestry and Medicine (ed. Tavantzis, S.) 165–190 (CRC, Boca Raton, 2002).
Jiang, D. & Ghabrial, S. A. Molecular characterization of Penicillium chrysogenum virus: reconsideration of the taxonomy of the genus Chrysovirus. J. Gen. Virol. 85, 2111–2121 (2004).
Acknowledgements
I am grateful to T. Allen, F. Deng, L. Geletka & G. Segers for suggestions and corrections. This work was supported in part by a Public Health Service grant.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Related links
DATABASES
Entrez Gene
FURTHER INFORMATION
Glossary
- BIOCONTROL
-
The use of one species of organism to eliminate or control another through a biological mechanism.
- ANASTOMOSIS
-
Fusion of somatic hyphae followed by exchange of cellular contents.
- TRANSGENIC HYPOVIRULENCE
-
Virulence-attenuated fungal strains that contain a chromosomally-integrated, infectious hypovirus cDNA.
- CONIDIA
-
Asexual spores.
- cDNA MICROARRAY
-
A collection of cDNA clones spotted on a support media such as a micoscope slide, in a high-density ordered array that is suitable for hybridization to appropriate nucleic-acid probes.
- EXPRESSED SEQUENCE TAG
-
(EST). A cloned cDNA copy of the mRNAs isolated from an organism. The combined collection (library) of EST clones represents the transcribed genes in that sample.
- VEGETATIVE INCOMPATIBILITY
-
A genetic system that controls the ability of two fungal strains to anastomose. In C. parasitica, this system consists of at least six genetic loci, each containing two alleles.
Rights and permissions
About this article
Cite this article
Nuss, D. Hypovirulence: Mycoviruses at the fungal–plant interface. Nat Rev Microbiol 3, 632–642 (2005). https://doi.org/10.1038/nrmicro1206
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1206
This article is cited by
-
Viruses of plant-pathogenic fungi: a promising biocontrol strategy for Sclerotinia sclerotiorum
Archives of Microbiology (2024)
-
Architecture of the dynamic fungal cell wall
Nature Reviews Microbiology (2023)
-
Podosphaera cerasi- an old foe of US sweet cherry with a new name –its biology, epidemiology, and beyond
Journal of Plant Pathology (2023)
-
Using some microorganisms as biocontrol agents to manage phytopathogenic fungi: a comprehensive review
Journal of Plant Pathology (2023)
-
The presence of mycoviral infection attenuates the growth and pathogenicity of the phytopathogenic fungus Botrytis cinerea collected from strawberry fields in Pakistan
European Journal of Plant Pathology (2023)